Publications from Dr. Snell and his group
Validating Metal Identities in 70 Metalloprotein Models with Particle-Induced X-ray Emission and X-ray Fluorescence Snell, EH. Grime, GW, Webb, SM, Costa, C., M. Snell, ME., Hunt, J., Tong, L., Montelione, GT., Zigweid, R., Staker, BL., Myler, PJ., and Garman, EF. Submitted.
A paper on the use of Particle Induced X-ray emission, and X-ray fluorescence Spectroscopy, to aid in metal identification and accurate structural modeling of metalloproteins.
Metals play essential structural and catalytical roles in biological mechanism yet many are modeled incorrectly. The scientific community utilizes these models often without knowledge of the potential errors. Using Particle Induced X-ray Emission (PIXE) and X-ray Fluorescence X-ray Spectroscopy (XRFS) to unambiguously identify the metals present, we examined the original protein samples used to submit 70 models to the Protein Data Bank (PDB), 28 from the North-East Structural Genomics consortium and 42 from the Seattle Structural Genomics Center for Infectious Diseases. The results were arranged into distinct classes based on the PIXE and XRFS measurements. The original model (OM) and new data were inconsistent in 40 (57%); extra metals not in the OM were detected in 29 (41%); and the OM metals and data agreed in one. In many of the 70 samples, problematic metal assignments can be identified from the difference electron density in the original structural model. The PIXE and XRFS results were in good agreement, confirming the potential for using high-throughput XRF more routinely to disambiguate metalloprotein identities. Our overall findings are a cause for concern, since PDB models are widely used by researchers unaware of the potential misidentification of metals. They are particularly worrying where machine learning is used to create artificial intelligence models to predict metal position, but more critically metal identity. On the positive side, the results demonstrate that metal identity can be rapidly determined, even from samples that have been prepared years before the measurement.
20 years of crystal hits: progress and promise in ultrahigh-throughput crystallization screening. Lynch, ML., Snell, ME., Potter, SA., Snell, EH., & Bowman. SEJ. Acta Crystallogr D Struct Biol. 2023 Mar 1;79(Pt 3):198-205.
A paper amply illustrating the complementary nature of cryo-EM and X-ray crystallography.
Diffraction-based structural methods contribute a large fraction of the biomolecular structural models available, providing a critical understanding of macromolecular architecture. These methods require crystallization of the target molecule, which remains a primary bottleneck in crystal-based structure determination. The National High-Throughput Crystallization Center at Hauptman–Woodward Medical Research Institute has focused on overcoming obstacles to crystallization through a combination of robotics-enabled high-throughput screening and advanced imaging to increase the success of finding crystallization conditions. This paper will describe the lessons learned from over 20 years of operation of our high-throughput crystallization services. The current experimental pipelines, instrumentation, imaging capabilities and software for image viewing and crystal scoring are detailed. New developments in the field and opportunities for further improvements in biomolecular crystallization are reflected on.
From Protein Design to the Energy Landscape of a Cold Unfolding Protein. Pulavarti SVSRK, Maguire JB, Yuen S, Harrison JS, Griffin J, Premkumar L, Esposito EA, Makhatadze GI, Garcia AE, Weiss TM, Snell EH, Kuhlman B, Szyperski T. J. Phys Chem B. 2022, 126, 6, 1212-1231.
Understanding protein folding is crucial for protein sciences. The conformational spaces and energy landscapes of cold (unfolded) protein states, as well as the associated transitions, are hardly explored. Furthermore, it is not known how structure relates to the cooperativity of cold transitions, if cold and heat unfolded states are thermodynamically similar, and if cold states play important roles for protein function. We created the cold unfolding 4-helix bundle DCUB1 with a de novo designed bipartite hydrophilic/hydrophobic core featuring a hydrogen bond network which extends across the bundle in order to study the relative importance of hydrophobic versus hydrophilic protein–water interactions for cold unfolding. Structural and thermodynamic characterization resulted in the discovery of a complex energy landscape for cold transitions, while the heat unfolded state is a random coil. Below ∼0 °C, the core of DCUB1 disintegrates in a largely cooperative manner, while a near-native helical content is retained. The resulting cold core-unfolded state is compact and features extensive internal dynamics. Below −5 °C, two additional cold transitions are seen, that is, (i) the formation of a water-mediated, compact, and highly dynamic dimer, and (ii) the onset of cold helix unfolding decoupled from cold core unfolding. Our results suggest that cold unfolding is initiated by the intrusion of water into the hydrophilic core network and that cooperativity can be tuned by varying the number of core hydrogen bond networks. Protein design has proven to be invaluable to explore the energy landscapes of cold states and to robustly test related theories.
Near-physiological-temperature serial crystallography reveals conformations of SARS-CoV-2 main protease active site for improved drug repurposing. Durdagi S, Dağ C, Dogan B, Yigin M, Avsar T, Buyukdag C, Erol I, Ertem FB, Calis S, Yildirim G, Orhan MD, Guven O, Aksoydan B, Destan E, Sahin K, Besler SO, Oktay L, Shafiei A, Tolu I, Ayan E, Yuksel B, Peksen AB, Gocenler O, Yucel AD, Can O, Ozabrahamyan S, Olkan A, Erdemoglu E, Aksit F, Tanisali G, Yefanov OM, Barty A, Tolstikova A, Ketawala GK, Botha S, Dao H, Hayes B, Liang M, Seaberg MH, Hunter MS, Batyuk A, Mariani V, Su Z, Poitevin F, Yoon CH, Kupitz C, Sierra RG, Snell EH, and DeMirci H. (2021) Structure 29, 12, 1382-1396.
The COVID-19 pandemic has resulted in 198 million reported infections and more than 4 million deaths as of July 2021. Research to identify effective therapies for COVID-19 includes: (1) designing a vaccine as future protection; (2) de novo drug discovery; and (3) identifying existing drugs to repurpose them as effective and immediate treatments. To assist in drug repurposing and design, we determine two apo structures of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) main protease at ambient temperature by serial femtosecond X-ray crystallography. We employ detailed molecular simulations of selected known main protease inhibitors with the structures and compare binding modes and energies. The combined structural and molecular modeling studies not only reveal the dynamics of small molecules targeting the main protease but also provide invaluable opportunities for drug repurposing and structure-based drug design strategies against SARS-CoV-2.
Biological networks across scales—The Theoretical and Empirical Foundations for Time-Varying Complex Networks that Connect Structure and Function across Levels of Biological Organization. Bogdan, P, Caetano-Anolles, Jolles, A, Kim, H, Morris, J, Murphy, C, Royer, Snell EH, Steinbrenner, A. and Strausfeld, N. (2021) Integrative and Comparative Biology 61, 6, 1991-2010.
A wide range of networks exist in biology, from gene regulatory networks important for organismal development, protein interaction networks that govern physiology and metabolism, and neural networks that store and convey information to networks of microbes that form microbiomes within hosts, animal contact networks that underlie social systems, and networks of populations on the landscape connected by migration. Increasing availability of extensive (big) data is amplifying our ability to quantify biological networks. Similarly, theoretical methods that describe network structure and dynamics are being developed. In this paper, we explore the opportunity for discovering universal laws connecting the structure of biological networks with their function, positioning them on the spectrum of time-evolving network structure, that is, dynamics of networks, from highly stable to exquisitely sensitive to perturbation. If such general laws exist, they could transform our ability to predict the response of biological systems to perturbations—an increasingly urgent priority in the face of anthropogenic changes to the environment that affect life across the gamut of organizational scales.
Structural biology in the time of COVID-19: perspectives on methods and milestones. Lynch, ML, Snell, EH, Bowman, SEJ. (2021) IUCrJ 8, 3, 335-341.
A review demonstrating the impact of complenentary techniques in structurally understanding the SARS-CoV-2 virus components.
The global COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) wreaked unprecedented havoc on global society. Yet the sheer magnitude and uniqueness of this event has also spawned a massive mobilization of effort in the scientific community to investigate the virus, to develop therapeutics and vaccines, and to understand the public health impacts. Structural biology has been at the center of these efforts, and we reflect on the status of structural science vis-a`-vis its role in the fight against COVID-19, to register the unprecedented response and to contemplate the role of structural biology in addressing future outbreak threats. Examining SARS-CoV-2 protein structures that have been deposited in the PDB since the virus was first identified allows a unique window into the power of structural biology and a snapshot of the advantages of the different techniques available, as well as insight into the complementarity of the structural methods.
A SAXS based approach to rationally evaluate radical scavengers – toward eliminating radiation damage in solution and crystallographic studies. Stachowski, TR, Snell, ME, Snell, EH. (2021) Journal of Synchrotron Radiation, 28, 1309-1320.
A sensitive method to evaluate free-radical scavengers in solution.
X-ray-based techniques are a powerful tool in structural biology but the radiation-induced chemistry that results can be detrimental and may mask an accurate structural understanding. Using a protein designed to undergo a large-scale structural change from breakage of a disulfide bond, radiation damage can be monitored with small-angle X-ray scattering. We have quantitatively evaluated how three scavengers commonly used in crystallographic experiments – sodium nitrate, cysteine, and ascorbic acid – perform in solution at 10°C. The SAXS-based method can detect damage at X-ray doses far lower than those accessible crystallographically, thereby providing a detailed picture of scavenger processes. Our engineered approach might provide a platform for more systematic and comprehensive screening of radioprotectants that can directly inform mitigation strategies for both solution and crystallographic experiments, while also clarifying fundamental radiation damage mechanisms and allowing the study of low dose radiation.
Microgravity as an environment for macromolecular crystallization – an outlook in the era of space stations and commercial space flight. Snell EH and Helliwell JR. (2021) Crystallography Reviews, 8th April, 1-44.
A review of the state of the field in microgravity based protein crystallization.
More regular space flight is becoming a reality, new diffraction data detectors have become available that have both a faster readout and lower noise, a new generation of extremely bright X-ray sources and X-ray free-electron lasers (XFELs) have become available with beam collimation properties well suited geometrically to more perfect protein crystals. Neutron sources, instrumentation, and methods have also advanced greatly for yielding complete structures at room temperature and radiation damage-free. The larger volumes of protein crystals from microgravity can synergise well with these recent neutron developments. Unfortunately, progress in harnessing these new technologies to maximize the benefits seen in microgravity-grown crystals has been patchy and even disappointing. Despite detailed theoretical analysis and key empirical studies, crystallization in microgravity has not yet produced the results that demonstrate its potential. In this review we present some of the key lessons learned and show how processes could yet be optimized given these new developments.
SAXS studies of X-ray induced disulfide bond damage: Engineering high-resolution insight from a low-resolution technique. Stachowski, T.R., Snell, M.E, and Snell, E.H. PLOS One (2020) 15(11): e0239702.
Developing a system to observe structural changes at very low X-ray doses.
Radition chemistry in biological X-ray crystallography iproduces both global and site-specific damage. Site specific damage can misdirect the biological interpretation of the structural models produced. The doses used for X-ray crystallography are typically in the kilo-gray to mega-gray range. While disulfide bonds are among the most significantly affected species in proteins in the crystalline state at both cryogenic and higher temperatures, there is limited information on their response to low X-ray doses in solution, the details of which might inform biomedical applications of X-rays. An engineered a protein approach enables small angle X-ray scattering (SAXS), to detect and monitor a residue specific process. The ability to generate a large-scale change through a site-specific interaction represents a promising tool for advancing radiation damage studies under solution conditions and in dose regimes of health-related impact.
Structural insights into conformational switching in latency-associated peptide between transforming growth factor β-1 bound and unbound states. Stachowski, T.R., Snell, M.E, & Snell, E.H. IUCrJ (2020) 7(2) 238-252.
Transforming growth factor -1 (TGF-1) is a secreted signalling protein that directs many cellular processes and is an attractive target for the treatment of several diseases. The primary endogenous activity regulatory mechanism for TGF-1 is sequestration by its pro-peptide, latency-associated peptide (LAP), which sterically prohibits receptor binding by caging TGF-1. As such, recombinant LAP is promising as a protein-based therapeutic for modulating TGF-1 activity; however, the mechanism of binding is incompletely understood. Comparison of the crystal structure of unbound LAP (solved here to 3.5 Å resolution) with that of the bound complex shows that LAP is in a more open and extended conformation when unbound to TGF-1. Analysis suggests a mechanism of binding TGF-1 through a large-scale conformational change that includes contraction of the inter-monomer interface and caging by the ‘straightjacket’ domain that may occur in partnership through a loop-to-helix transition in the core jelly-roll fold. This conformational change does not appear to include a repositioning of the integrin-binding motif as previously proposed. X-ray scattering-based modelling supports this mechanism and reveals possible orientations and ensembles in solution. Although native LAP is heavily glycosylated, solution scattering experiments show that the overall folding and flexibility of unbound LAP are not influenced by glycan modification. The combination of crystallography, solution scattering and biochemical experiments reported here provide insight into the mechanism of LAP sequestration of TGF-1 that is of fundamental importance for therapeutic development.
High-Throughput PIXE as an Essential Quantitative Assay for Accurate Metalloprotein Structural Analysis: Development and Application. Grime, G.W., Zeldin, O.B., Snell, M.E., Lowe, E.D., Hunt, J.F., Montelione, G.T., Tong, L., Snell, E.H.* & Garman, E.F* – joint corresponding. JACS (2020) 142, 185-197.
Revealing the misidentification of metals in the PDB and the new information resulting when identity is corrected.
Metalloproteins comprise over one-third of proteins, with approximately half of all enzymes requiring metal to function. Accurate identification of these metal atoms and their environment is a prerequisite to understanding biological mechanism. Using ion beam analysis through particle induced X-ray emission (PIXE), we have quantitatively identified the metal atoms in 30 previously structurally characterized proteins using minimal sample volume and a high-throughput approach. Over half of these metals had been misidentified in the deposited structural models. Some of the PIXE detected metals not seen in the models were explainable as artifacts from promiscuous crystallization reagents. For others, using the correct metal improved the structural models. For multinuclear sites, anomalous diffraction signals enabled the positioning of the correct metals to reveal previously obscured biological information. PIXE is insensitive to the chemical environment, but coupled with experimental diffraction data deposited alongside the structural model it enables validation and potential remediation of metalloprotein models, improving structural and, more importantly, mechanistic knowledge.
Structural consequences of transforming growth factor beta-1 activation from near-therapeutic X-ray doses, Stachowski, T., Grant, T.D. and Snell, E.H. Journal of Synchrotron radiation (2019), 26, 967-979.
Dissociation of transforming growth factor beta-1 (TGFβ-1) from the inhibitory protein latency-associated peptide (LAP) can occur from low doses of X-ray irradiation of the LAP–TGFβ-1 complex, resulting in the activation of TGFβ-1, and can have health-related consequences. Using the tools and knowledge developed in the study of radiation damage in the crystallographic setting, small-angle X-ray scattering (SAXS) and complementary techniques suggest an activation process that is initiated but not driven by the initial X-ray exposure. LAP is revealed to be extended when not bound to TGFβ-1 and has a different structural conformation compared to the bound state. These studies pave the way for the structural understanding of systems impacted at therapeutic X-ray doses and show the potential impact of radiation damage studies beyond their original intent.
Structural knowledge or X-ray damage. A case study on xylose isomerase illustrating both. Taberman, H, Bury, CS, van der Woerd MJ, Snell EH, Garman EF. J. Synchrotron Radiation. (2019). 26, 931-944.
Showing how radiation damage artifacts can be mechanistically interpreted.
Xylose isomerase (XI) is an industrially important metalloprotein studied for decades. Its reaction mechanism has been postulated to involve movement of the catalytic metal cofactor to several different conformations. Here, a dosedependent approach was used to investigate the radiation damage effects on XI and their potential influence on the reaction mechanism interpreted from the X-ray derived structures. Radiation damage is still one of the major challenges for X-ray diffraction experiments and causes both global and site-specific damage. In this study, consecutive high-resolution data sets from a single XI crystal from the same wedge were collected at 100 K and the progression of radiation damage was tracked over increasing dose (0.13–3.88 MGy). The catalytic metal and its surrounding amino acid environment experience a buildup of free radicals, and the results show radiation-damage-induced structural perturbations ranging from an absolute metal positional shift to specific residue motions in the active site. The apparent metal movement is an artefact of global damage and the resulting unit-cell expansion, but residue motion appears to be driven by the dose. Understanding and identifying radiation-induced damage is an important factor in accurately interpreting the biological conclusions being drawn.
Protein and RNA dynamical fingerprinting. Niessen KA, Xu M, George DK, Chen MC, Ferré-D’Amaré AR, Snell EH, Cody V, Pace J, Schmidt M, Markelz AG.Nat Commun. 2019 Mar 4;10(1):1026.
Protein structural vibrations impact biology by steering the structure to functional intermediate states; enhancing tunneling events; and optimizing energy transfer. Strong water absorption and a broad continuous vibrational density of states have prevented optical identification of these vibrations. Recently spectroscopic signatures that change with functional state were measured using anisotropic terahertz microscopy. The technique however has complex sample positioning requirements and long measurement times, limiting access for the biomolecular community. Here we demonstrate that a simplified system increases spectroscopic structure to dynamically fingerprint biomacromolecules with a factor of 6 reduction in data acquisition time. Using this technique, polarization varying anisotropy terahertz microscopy, we show sensitivity to inhibitor binding and unique vibrational spectra for several proteins and an RNA G-quadruplex. The technique’s sensitivity to anisotropic absorbance and birefringence provides rapid assessment of macromolecular dynamics that impact biology.
Classification of crystallization outcomes using deep convolutional neural networks. Bruno AE, Charbonneau, P, Newman J, Snell EH, So DR, Vanhoucke V, Watkins CJ, Williams S, Wilson J. PLoS ONE 2018 13(6):e0198883.
Establishing the first reliable crystallization outcome classifier with a performance better than a human.
The Machine Recognition of Crystallization Outcomes (MARCO) initiative has assembled roughly half a million annotated images of macromolecular crystallization experiments from various sources and setups. Here, state-of-the-art machine learning algorithms are trained and tested on different parts of this data set. We find that more than 94% of the test images can be correctly labeled, irrespective of their experimental origin. Because crystal recognition is key to high-density screening and the systematic analysis of crystallization experiments, this approach opens the door to both industrial and fundamental research applications.
Biological Small Angle Scattering Theory and Practice. Lattman, EE., Grant, TD, Snell, EH. Oxford University Press. 2018.
A book aimed at crystallographers and other structural biologists to introduce them to SAX/SANS techniques, provide practical guidance, and introduce the information that can be obtained from them.
Small angle solution scattering (SAS) is increasingly being applied to biological problems. It is a complementary technique that, when applied in appropriate circumstances with carefully structured questions, can provide unique information not available from other techniques. While small angle solution scattering has been around for some time, a confluence of recent developments has dramatically enhanced its power. Intense third generation X-ray sources, low noise detectors, development of new algorithms and the computational power to take advantage of these have all matured, and use of free-electron x-ray laser sources is on the horizon. Whole new classes of experiments and analyses have been created as a result. These include the generation of molecular envelopes, the ability to do time-resolved studies, and the ability to account for structural changes using modelling based on the SAS data. The technical improvements have also reduced the amount of time and material needed to carry out an experiment. Beamtime at synchrotron sources is in demand, workshops on the subject are popular and researchers adopting the technique as part of their repertoire are growing. With these in mind, this book was written to guide structural biologists who may wish to adopt the technique, understand its strengths and weaknesses or just have a general interest in its potential.
Double-flow focused liquid injector for efficient serial femtosecond crystallography. Oberthuer D, Knoška J, Wiedorn MO, Beyerlein KR, Bushnell DA, Kovaleva EG, Heymann M, Gumprecht L, Kirian RA, Barty A, Mariani V, Tolstikova A, Adriano L, Awel S, Barthelmess M, Dörner K, Xavier PL, Yefanov O, James DR, Nelson G, Wang D, Calvey G, Chen Y, Schmidt A, Szczepek M, Frielingsdorf S, Lenz O, Snell E, Robinson PJ, Šarler B, Belšak G, Maček M, Wilde F, Aquila A, Boutet S, Liang M, Hunter MS, Scheerer P, Lipscomb JD, Weierstall U, Kornberg RD, Spence JC, Pollack L, Chapman HN, Bajt S. Sci Rep. (2017) Mar 16;7:44628.
Serial femtosecond crystallography requires reliable and efficient delivery of fresh crystals across the beam of an X-ray free-electron laser over the course of an experiment. We introduce a double-flow focusing nozzle to meet this challenge, with significantly reduced sample consumption, while improving jet stability over previous generations of nozzles. We demonstrate its use to determine the first room-temperature structure of RNA polymerase II at high resolution, revealing new structural details. Moreover, the double-flow focusing nozzles were successfully tested with three other protein samples and the first room temperature structure of an extradiol ring-cleaving dioxygenase was solved by utilizing the improved operation and characteristics of these devices.
Moving in the Right Direction: Protein Vibrational Steering Function. Niessen KA, Xu M, Paciaroni A, Orecchini A, Snell EH, Markelz AG. Biophys J. 2017;112:933-942.
Nearly all protein functions require structural change, such as enzymes clamping onto substrates, and ion channels opening and closing. These motions are a target for possible new therapies; however, the control mechanisms are under debate. Calculations have indicated protein vibrations enable structural change. However, previous measurements found these vibrations only weakly depend on the functional state. By using the novel technique of anisotropic terahertz microscopy, we find that there is a dramatic change to the vibrational directionality with inhibitor binding to lysozyme, whereas the vibrational energy distribution, as measured by neutron inelastic scattering, is only slightly altered. The anisotropic terahertz measurements provide unique access to the directionality of the intramolecular vibrations, and immediately resolve the inconsistency between calculations and previous measurements, which were only sensitive to the energy distribution. The biological importance of the vibrational directions versus the energy distribution is revealed by our calculations comparing wild-type lysozyme with a higher catalytic rate double deletion mutant. The vibrational energy distribution is identical, but the more efficient mutant shows an obvious reorientation of motions. These results show that it is essential to characterize the directionality of motion to understand and control protein dynamics to optimize or inhibit function.
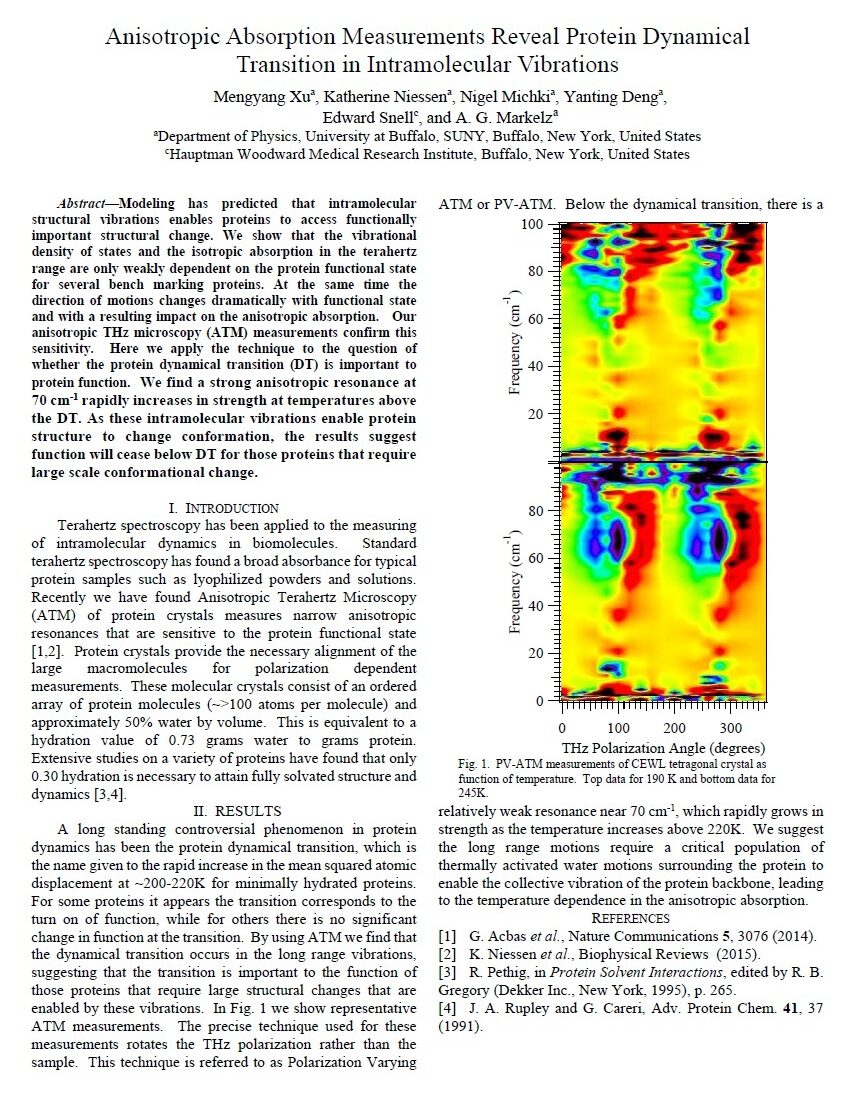
Anisotropic absorption measurements reveal protein dynamical transition in intramolecular vibrations, M. Xu, K. Niessen, N. Michki, Y. Deng, E. Snell and A. G. Markelz, 2016 41st International Conference on Infrared, Millimeter, and Terahertz waves (IRMMW-THz), Copenhagen, Denmark, 2016, pp. 1-1
Modeling has predicted that intramolecular structural vibrations enables proteins to access functionally important structural change. We show that the vibrational density of states and the isotropic absorption in the terahertz range are only weakly dependent on the protein functional state for several bench marking proteins. At the same time the direction of motions changes dramatically with functional state and with a resulting impact on the anisotropic absorption. Our anisotropic THz microscopy (ATM) measurements confirm this sensitivity. Here we apply the technique to the question of whether the protein dynamical transition (DT) is important to protein function. We find a strong anisotropic resonance at 70 cm-1 rapidly increases in strength at temperatures above the DT. As these intramolecular vibrations enable protein structure to change conformation, the results suggest function will cease below DT for those proteins that require large scale conformational change.
The use of haptic interfaces and web services in crystallography: an application for a `screen to beam’ interface. Bruno A, Soares A, Owen R, Snell EH. J. Appl. Cryst. (2016). 49, 2082-2090.
Developing user-friendly interfaces for the gene to structure pipeline and experiences with data collection after shipping crystals at room temperature across continents (across the pond).
Haptic interfaces have become common in consumer electronics. They enable easy interaction and information entry without the use of a mouse or keyboard. The work presented here illustrates the application of a haptic interface to crystallization screening in order to provide a natural means for visualizing and selecting results. By linking this to a cloud-based database and web-based application program interface, the same application shifts the approach from ‘point and click’ to ‘touch and share’, where results can be selected, annotated and discussed collaboratively. In the crystallographic application, given a suitable crystallization plate, beamline and robotic end effector, the resulting information can be used to close the loop between screening and X-ray analysis, allowing a direct and efficient ‘screen to beam’ approach. The application is not limited to the area of crystallization screening; ‘touch and share’ can be used by any information-rich scientific analysis and geographically distributed collaboration
Computational crystallization. Altan I, Charbonneau P, Snell EH. Arch Biochem Biophys. 2016 Jan 11. pii: S0003-9861(16)30004-2.
Crystallization is a key step in macromolecular structure determination by crystallography. While a robust theoretical treatment of the process is available, due to the complexity of the system, the experimental process is still largely one of trial and error. In this article, efforts in the field are discussed together with a theoretical underpinning using a solubility phase diagram. Prior knowledge has been used to develop tools that computationally predict the crystallization outcome and define mutational approaches that enhance the likelihood of crystallization. For the most part these tools are based on binary outcomes (crystal or no crystal), and the full information contained in an assembly of crystallization screening experiments is lost. The potential of this additional information is illustrated by examples where new biological knowledge can be obtained and where a target can be sub-categorized to predict which class of reagents provides the crystallization driving force. Computational analysis of crystallization requires complete and correctly formatted data. While massive crystallization screening efforts are under way, the data available from many of these studies are sparse. The potential for this data and the steps needed to realize this potential are discussed.
The detection and subsequent volume optimization of biological nanocrystals, Luft JR, Wolfley JR, Franks EC, Lauricella AM, Gualtieri EJ, Snell EH, Xiao R, Everett JK, Montelione GT. Struct Dyn. 2015 May 15;2(4):041710.
A study on producing crystals of defined volume.
Identifying and then optimizing initial crystallization conditions is a prerequisite for macromolecular structure determination by crystallography. Improved technologies enable data collection on crystals that are difficult if not impossible to detect using visible imaging. The application of second-order nonlinear imaging of chiral crystals and ultraviolet two-photon excited fluorescence detection is shown to be applicable in a high-throughput manner to rapidly verify the presence of
nanocrystals in crystallization screening conditions. It is noted that the nanocrystals are rarely seen without also producing microcrystals from other chemical conditions. A crystal volume optimization method is described and associated with a phase diagram for crystallization.
The structure of the PanD/PanZ protein complex reveals negative feedback regulation of pantothenate biosynthesis by coenzyme A. Monteiro DC, Patel V, Bartlett CP, Nozaki S, Grant TD, Gowdy JA, Thompson GS, Kalverda AP, Snell EH, Niki H, Pearson AR, Webb ME. Chem Biol. 2015 Apr 23;22(4):492-503
Coenzyme A (CoA) is an ubiquitous and essential cofactor, synthesized from the precursor pantothenate. Vitamin biosynthetic pathways are normally tightly regulated, including the pathway from pantothenate to CoA. However, no regulation of pantothenate biosynthesis has been identified. We have recently described an additional component in the pantothenate biosynthetic pathway, PanZ, which promotes the activation of the zymogen, PanD, to form aspartate a-decarboxylase (ADC) in a CoA dependent manner. Here we report the structure of PanZ in complex with PanD, which reveals the structural basis for the CoA dependence of this interaction and activation. In addition, we show that PanZ acts as a CoA-dependent inhibitor of ADC catalysis. This inhibitory effect can effectively regulate the biosynthetic pathway to pantothenate, and thereby also regulate CoA biosynthesis. This represents a previously unobserved mode of metabolic regulation whereby a cofactor-utilizing protein negatively regulates the biosynthesis of the same cofactor.
The accurate assessment of small-angle X-ray scattering data. Grant TD, Luft JR, Carter LG, Matsui T, Weiss TM, Martel A, Snell EH. Acta Crystallogr D Biol Crystallogr. 2015 Jan 1;71(Pt 1):45-56.
Metrics to evaluate the quality of SAXS data.
Small-angle X-ray scattering (SAXS) has grown in popularity in recent times with the advent of bright synchrotron X-ray sources, powerful computational resources and algorithms enabling the calculation of increasingly complex models. However, the lack of standardized data-quality metrics presents difficulties for the growing user community in accurately assessing the quality of experimental SAXS data. Here, a series of metrics to quantitatively describe SAXS data in an objective manner using statistical evaluations are defined. These metrics are applied to identify the effects of radiation damage, concentration dependence and interparticle interactions on SAXS data from a set of 27 previously described targets for which high-resolution structures have been determined via X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy. The studies show that these metrics are sufficient to characterize SAXS data quality on a small sample set with statistical rigor and sensitivity similar to or better than manual analysis. The development of data-quality analysis strategies such as these initial efforts is needed to enable the accurate and unbiased assessment of SAXS data quality
A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta. Rossi P, Shi L, Liu G, Barbieri CM, Lee HW, Grant TD, Luft JR, Xiao R, Acton TB, Snell EH, Montelione GT, Baker D, Lange OF, Sgourakis NG. Proteins. 2015 Feb;83(2):309-17.
Oligomeric proteins are important targets for structure determination in solution. While in most cases the fold of individual subunits can be determined experimentally, or predicted by homology-based methods, protein–protein interfaces are challenging to determine de novo using conventional NMR structure determination protocols. Here we focus on a member of the bet-V1 superfamily, Aha1 from Colwellia psychrerythraea. This family displays a broad range of crystallographic interfaces none of which can be reconciled with the NMR and SAXS data collected for Aha1. Unlike conventional methods relying on a dense network of experimental restraints, the sparse data are used to limit conformational search during optimization of a physically realistic energy function. This work highlights a new approach for studying minor conformational changes due to structural plasticity within a single dimeric interface in solution.
Comparing chemistry to outcome: the development of a chemical distance metric, coupled with clustering and hierarchal visualization applied to macromolecular crystallography. Bruno AE, Ruby AM, Luft JR, Grant TD, Seetharaman J, Montelione GT, Hunt JF, Snell EH. PLoS One. 2014 Jun 27;9(6):e100782.
Analyzing crystallization outcome as a whole rather than individually and producing new mechanistic information.
Many bioscience fields employ high-throughput methods to screen multiple biochemical conditions. The analysis of these becomes tedious without a degree of automation. Crystallization, a rate limiting step in biological X-ray crystallography, is one of these fields. Screening of multiple potential crystallization conditions (cocktails) is the most effective method of probing a proteins phase diagram and guiding crystallization but the interpretation of results can be time-consuming. To aid this empirical approach a cocktail distance coefficient was developed to quantitatively compare macromolecule crystallization conditions and outcome. These coefficients were evaluated against an existing similarity metric developed for crystallization, the C6 metric, using both virtual crystallization screens and by comparison of two related 1,536-cocktail highthroughput crystallization screens. Hierarchical clustering was employed to visualize one of these screens and the crystallization results from an exopolyphosphatase-related protein from Bacteroides fragilis, (BfR192) overlaid on this clustering. This demonstrated a strong correlation between certain chemically related clusters and crystal lead conditions. While this analysis was not used to guide the initial crystallization optimization, it led to the re-evaluation of unexplained peaks in the electron density map of the protein and to the insertion and correct placement of sodium, potassium and phosphate atoms in the structure. With these in place, the resulting structure of the putative active site demonstrated features consistent with active sites of other phosphatases which are involved in binding the phosphoryl moieties of nucleotide triphosphates. The new distance coefficient, CDcoeff, appears to be robust in this application, and coupled with hierarchical clustering and the overlay of crystallization outcome, reveals information of biological relevance. While tested with a single example the potential applications related to crystallography appear promising and the distance coefficient, clustering, and hierarchal visualization of results undoubtedly have applications in wider fields.
Identifying, studying and making good use of macromolecular crystals. Calero G, Cohen AE, Luft JR, Newman J, Snell EH.Acta Crystallogr F Struct Biol Commun. 2014 Aug;70(Pt 8):993-1008.
A review on the methods to identify and make good use of crystals that result. Part of the IUCr Crystallization Series.
Structural biology has contributed tremendous knowledge to the understanding of life on the molecular scale. The Protein Data Bank, a depository of this structural knowledge, currently contains over 100 000 protein structures, with the majority stemming from X-ray crystallography. As the name might suggest, crystallography requires crystals. As detectors become more sensitive and X-ray sources more intense, the notion of a crystal is gradually changing from one large enough to embellish expensive jewellery to objects that have external dimensions of the order of the wavelength of visible light. Identifying these crystals is a prerequisite to their study. This paper discusses developments in identifying these crystals during crystallization screening and distinguishing them from other potential outcomes. The practical aspects of ensuring that once a crystal is identified it can then be positioned in the X-ray beam for data collection are also addressed.
Crystallization screening: the influence of history on current practice. Luft JR, Newman J, Snell EH. Acta Crystallogr F Struct Biol Commun. 2014 Jul;70(Pt 7):835-53.
A history of crystallization screen development with reference to the Crystallization Screening Center. Part of the IUCr Crystallization Series.
While crystallization historically predates crystallography, it is a critical step for the crystallographic process. The rich history of crystallization and how that history influences current practices is described. The tremendous impact of crystallization screens on the field is discussed.
Statistical analysis of crystallization database links protein physico-chemical features with crystallization mechanisms. Fusco D, Barnum TJ, Bruno AE, Luft JR, Snell EH, Mukherjee S, Charbonneau P. PLoS One. 2014 Jul 2;9(7):e101123.
Using the Crystallization Screening Center results fromt he Protein Structure Initiative to glean information about crystallization approaches.
X-ray crystallography is the predominant method for obtaining atomic-scale information about biological macromolecules. Despite the success of the technique, obtaining well diffracting crystals still critically limits going from protein to structure. In practice, the crystallization process proceeds through knowledge-informed empiricism. Better physico-chemical understanding remains elusive because of the large number of variables involved, hence little guidance is available to systematically identify solution conditions that promote crystallization. To help determine relationships between macromolecular properties and their crystallization propensity, we have trained statistical models on samples for 182 proteins supplied by the Northeast Structural Genomics consortium. Gaussian processes, which capture trends beyond the reach of linear statistical models, distinguish between two main physico-chemical mechanisms driving crystallization. One is characterized by low levels of side chain entropy and has been extensively reported in the literature. The other identifies specific electrostatic interactions not previously described in the crystallization context. Because evidence for two distinct mechanisms can be gleaned both from crystal contacts and from solution conditions leading to successful crystallization, the model offers future avenues for optimizing crystallization screens based on partial structural information. The availability of crystallization data coupled with structural outcomes analyzed through state-of-the-art statistical models may thus guide macromolecular crystallization toward a more rational basis.
A new view on crystal harvesting. Luft JR, Grant TD, Wolfley JR, Snell EH. J Appl Crystallogr. 2014 May 29;47(Pt 3):1158-1161.
Deconflicting the observation pathway from the manipulation pathway.
X-ray crystallography typically requires the mounting of crystals, which can make the sample difficult to manipulate when it is small and the microscope objective is close to the crystallization plate. By simply moving the objective to the bottom of a clear crystallization plate (inverting the normal view), crystals were able to be manipulated and harvested from wells having a 0.9 mm diameter and 5.0 mm depth. The mounting system enabled the structural solution of the 187 amino acid N-terminal domain of Saccharomyces cerevisiae glutaminyltRNA synthetase from crystals that appeared during high-throughput screening but proved recalcitrant to scale-up and optimization. While not a general mounting solution, the simple expedient of removing the objective lens from the area where manipulation and harvesting occur greatly facilitates the manual, or even automated, process.
Optical measurements of long-range protein vibrations. Acbas G, Niessen KA, Snell EH, Markelz AG. Nat Commun. 2014;5:3076.
Protein biological function depends on structural flexibility and change. From cellular communication through membrane ion channels to oxygen uptake and delivery by haemoglobin, structural changes are critical. It has been suggested that vibrations that extend through the protein play a crucial role in controlling these structural changes. While nature may utilize such long-range vibrations for optimization of biological processes, bench-top characterization of these extended structural motions for engineered biochemistry has been elusive. Here we show the first optical observation of long-range protein vibrational modes. This is achieved by orientation-sensitive terahertz near-field microscopy measurements of chicken egg white lysozyme single crystals. Underdamped modes are found to exist for frequencies >10 cm^-1. The existence of these persisting motions indicates that damping and intermode coupling are weaker than previously assumed. The methodology developed permits protein engineering based on dynamical network optimization.
Neutron structure of the cyclic glucose-bound xylose isomerase E186Q mutant. Munshi P, Snell EH, van der Woerd MJ, Judge RA, Myles DA, Ren Z, Meilleur F. Acta Crystallogr D Biol Crystallogr. 2014 Feb;70(Pt 2):414-20.
Ketol-isomerases catalyze the reversible isomerization between aldoses and ketoses. d-Xylose isomerase carries out the first reaction in the catabolism of d-xylose, but is also able to convert d-glucose to d-fructose. The first step of the reaction is an enzyme-catalyzed ring opening of the cyclic substrate. The active-site amino-acid acid/base pair involved in ring opening has long been investigated and several models have been proposed. Here, the structure of the xylose isomerase E186Q mutant with cyclic glucose bound at the active site, refined against joint X-ray and neutron diffraction data, is reported. Detailed analysis of the hydrogen-bond networks at the active site of the enzyme suggests that His54, which is doubly protonated, is poised to protonate the glucose O5 position, while Lys289, which is neutral, promotes deprotonation of the glucose O1H hydroxyl group via an activated water molecule. The structure also reveals an extended hydrogen-bonding network that connects the conserved residues Lys289 and Lys183 through three structurally conserved water molecules and residue 186, which is a glutamic acid to glutamine mutation.
Measuring phonons in protein crystals. Acbas G, Niessen KA, George, DK, Snell E, Markelz AG. Ultrafast Phenomena and Nanophotonics XVII, Proc. of SPIE, 2013, Vol. 8623, 862305, 1-4.
Using Terahertz near field microscopy we find orientation dependent narrow band absorption features for lysozyme crystals. Here we discuss identification of protein collective modes associated with the observed features. Using normal mode calculations we find good agreement with several of the measured features, suggesting that the modes arise from internal molecular motions and not crystal phonons. Such internal modes have been associated with protein function.
The structure of yeast glutaminyl-tRNA synthetase and modeling of its interaction with tRNA. Grant TD, Luft JR, Wolfley JR, Snell ME, Tsuruta H, Corretore S, Quartley E, Phizicky EM, Grayhack EJ, Snell EH. J Mol Biol. 2013 Jul 24;425(14):2480-93.
Eukaryotic glutaminyl-tRNA synthetase (GlnRS) contains an appended N-terminal domain (NTD) whose precise function is unknown. Although GlnRS structures from two prokaryotic species are known, no eukaryotic GlnRS structure has been reported. Here we present the first crystallographic structure of yeast GlnRS, finding that the structure of the C-terminal domain is highly similar to Escherichia coli GlnRS but that 214 residues, including the NTD, are crystallographically disordered. We present a model of the full-length enzyme in solution, using the structures of the C-terminal domain, and the isolated NTD, with small-angle X-ray scattering data of the full-length molecule. We proceed to model the enzyme bound to tRNA, using the crystallographic structures of GatCAB and GlnRS–tRNA complex from bacteria. We contrast the tRNA-bound model with the tRNA-free solution state and perform molecular dynamics on the full-length GlnRS–tRNA complex, which suggests that tRNA binding involves the motion of a conserved hinge in the NTD.
Purification and SAXS analysis of the integrin linked kinase, PINCH, parvin (IPP) heterotrimeric complex. Stiegler AL, Grant TD, Luft JR, Calderwood DA, Snell EH, Boggon TJ. PLoS One. 2013;8(1):e55591.
The heterotrimeric protein complex containing the integrin linked kinase (ILK), parvin, and PINCH proteins, termed the IPP complex, is an essential component of focal adhesions, where it interacts with many proteins to mediate signaling from integrin adhesion receptors. Here we conduct a biochemical and structural analysis of the minimal IPP complex, comprising full-length human ILK, the LIM1 domain of PINCH1, and the CH2 domain of a-parvin. We provide a detailed purification protocol for IPP and show that the purified IPP complex is stable and monodisperse in solution. Using small-angle X-ray scattering (SAXS), we also conduct the first structural characterization of IPP, which reveals an elongated shape with dimensions 120660640 A˚. Flexibility analysis using the ensemble optimization method (EOM) is consistent with an IPP complex structure with limited flexibility, raising the possibility that inter-domain interactions exist. However, our studies suggest that the inter-domain linker in ILK is accessible and we detect no inter-domain contacts by gel filtration analysis. This study provides a structural foundation to understand the conformational restraints that govern the IPP complex.
Insights into the mechanism of X-ray-induced disulfide-bond cleavage in lysozyme crystals based on EPR, optical absorption and X-ray diffraction studies. Sutton KA, Black PJ, Mercer KR, Garman EF, Owen RL, Snell EH, Bernhard WA. Acta Crystallogr D Biol Crystallogr. 2013 Dec;69(Pt 12):2381-94.
Understanding the radiation chemistry pathway for disulfide bon damage.
Electron paramagnetic resonance (EPR) and online UV–visible absorption microspectrophotometry with X-ray crystallography have been used in a complementary manner to follow X-ray-induced disulfide-bond cleavage. Online UV– visible spectroscopy showed that upon X-irradiation, disulfide radicalization appeared to saturate at an absorbed dose of approximately 0.5–0.8 MGy, in contrast to the saturating dose of 0.2 MGy observed using EPR at much lower dose rates. The observations suggest that a multi-track model involving product formation owing to the interaction of two separate tracks is a valid model for radiation damage in protein crystals. The saturation levels are remarkably consistent given the widely different experimental parameters and the range of total absorbed doses studied. The results indicate that even at the lowest doses used for structural investigations disulfide bonds are already radicalized. Multi-track considerations offer the first step in a comprehensive model of radiation damage that could potentially lead to a combined computational and experimental approach to identifying when damage is likely to be present, to quantitate it and to provide the ability to recover the native unperturbed structure.
On the need for an international effort to capture, share and use crystallization screening data. Newman J, Bolton EE, Müller-Dieckmann J, Fazio VJ, Gallagher DT, Lovell D, Luft JR, Peat TS, Ratcliffe D, Sayle RA, Snell EH, Taylor K, Vallotton P, Velanker S, von Delft F. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2012 Mar 1;68(Pt 3):253-8.
A call for standards in crystallization chemical descriptions therby enabling efficient computational mining of data.
When crystallization screening is conducted many outcomes are observed but typically the only trial recorded in the literature is the condition that yielded the crystal(s) used for subsequent diffraction studies. The initial hit that was optimized and the results of all the other trials are lost. These missing results contain information that would be useful for an improved general understanding of crystallization. This paper provides a report of a crystallization data exchange (XDX) workshop organized by several international large-scale crystallization
screening laboratories to discuss how this information may be captured and utilized. A group that administers a significant fraction of the world’s crystallization screening results was convened, together with chemical and structural data informaticians and computational scientists who specialize in creating and analysing large disparate data sets. The development of a crystallization ontology for the crystallization community was proposed. This paper (by the attendees of the workshop) provides the thoughts and rationale leading to this conclusion. This is brought to the attention of the wider audience of crystallographers so that they are aware of these early efforts and can contribute to the process going forward.
Structural conservation of an ancient tRNA sensor in eukaryotic glutaminyl-tRNA synthetase. Grant TD, Snell EH, Luft JR, Quartley E, Corretore S, Wolfley JR, Snell ME, Hadd A, Perona JJ, Phizicky EM, Grayhack EJ. Nucleic Acids Res. 2012 Apr;40(8):3723-31.
In all organisms, aminoacyl tRNA synthetases covalently attach amino acids to their cognate tRNAs. Many eukaryotic tRNA synthetases have acquired appended domains, whose origin, structure and function are poorly understood. The N-terminal appended domain (NTD) of glutaminyl-tRNA synthetase (GlnRS) is intriguing since GlnRS is primarily a eukaryotic enzyme, whereas in other kingdoms Gln-tRNAGln is primarily synthesized by first forming Glu-tRNAGln, followed by conversion to Gln-tRNAGln by a tRNA-dependent amidotransferase. We report a functional and structural analysis of the NTD of Saccharomyces cerevisiae GlnRS, Gln4. Yeast mutants lacking the NTD exhibit growth defects, and Gln4 lacking the NTD has reduced complementarity for tRNAGln and glutamine. The 187-amino acid Gln4 NTD, crystallized and solved at 2.3 A˚ resolution, consists of two subdomains, each exhibiting an extraordinary structural resemblance to adjacent tRNA specificitydetermining domains in the GatB subunit of the GatCAB amidotransferase, which forms Gln-tRNAGln. These subdomains are connected by an apparent hinge comprised of conserved residues. Mutation of these amino acids produces Gln4 variants with reduced affinity for tRNAGln, consistent with a hinge-closing mechanism proposed for GatB recognition of tRNA. Our results suggest a possible origin and function of the NTD that would link the phylogenetically diverse mechanisms of Gln-tRNAGln synthesis.
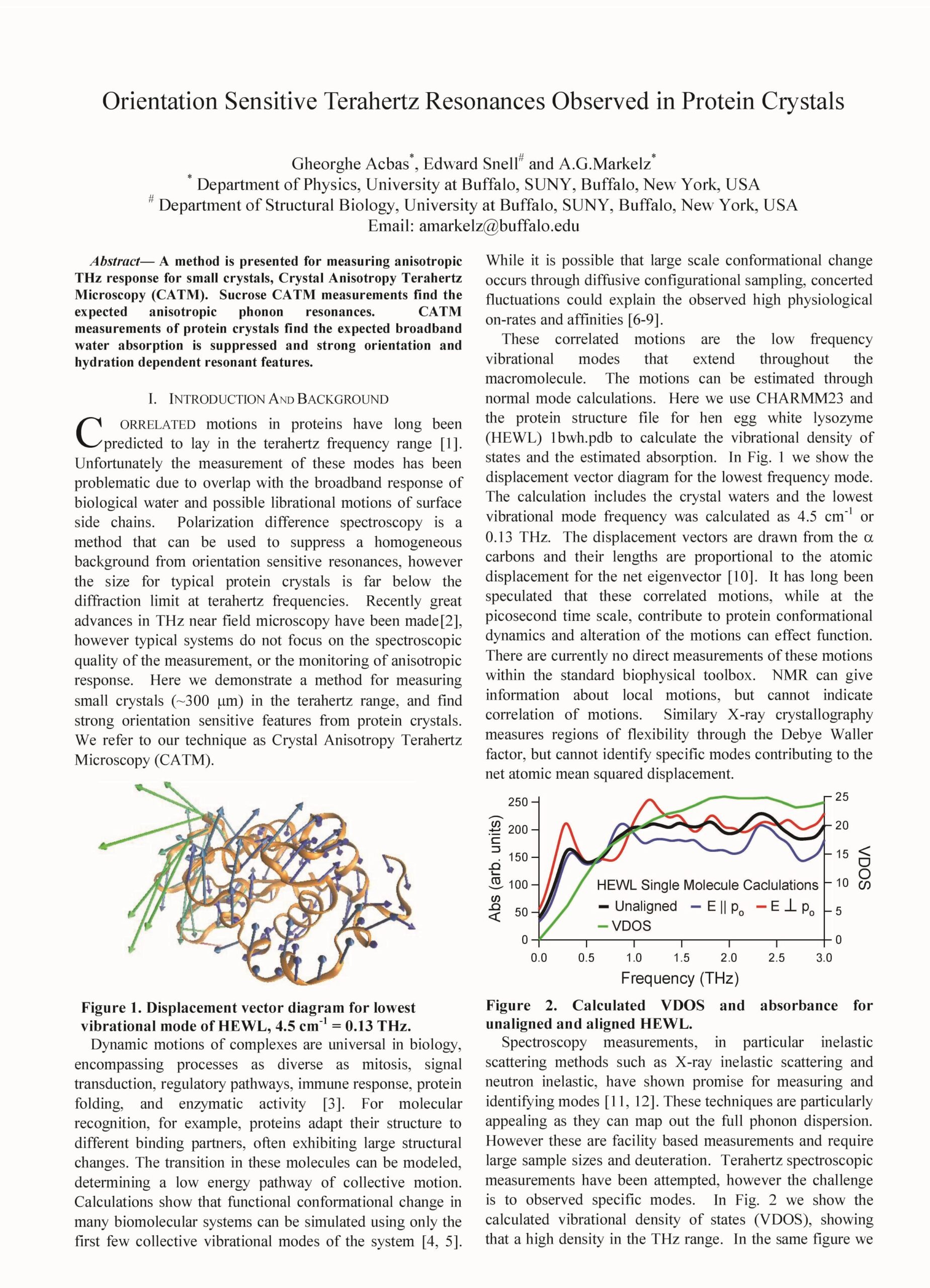
Orientation sensitive Terahertz resonances observed in protein crystals, G. Acbas, E. Snell and A. G. Markelz, 2012 37th International Conference on Infrared, Millimeter, and Terahertz Waves, Wollongong, NSW, Australia, 2012, pp. 1-3
A method is presented for measuring anisotropic THz response for small crystals, Crystal Anisotropy Terahertz Microscopy (CATM). Sucrose CATM measurements find the expected anisotropic phonon resonances. CATM measurements of protein crystals find the expected broadband water absorption is suppressed and strong orientation and hydration dependent resonant features.
What’s in a drop? Correlating observations and outcomes to guide macromolecular crystallization experiments. Luft JR, Wolfley JR, Snell EH. Cryst Growth Des. 2011 Mar 2;11(3):651-663.
Explaining many of the outcomes of crystallization experiments and how combined, they guide optimization and success.
Observations of crystallization experiments are classified as specific outcomes and integrated through a phase diagram to visualize solubility and thereby direct subsequent experiments. Specific examples are taken from our highthroughput crystallization laboratory which provided a broad scope of data from 20 million crystallization experiments on 12 500 different biological macromolecules. The methods and rationale are broadly and generally applicable in any crystallization laboratory. Through a combination of incomplete factorial sampling of crystallization cocktails, standard outcome classifications, visualization of outcomes as they relate chemically, and application of a simple phase diagram approach, we demonstrate how to logically design subsequent crystallization experiments.
Lessons from high-throughput protein crystallization screening: 10 years of practical experience. Luft JR, Snell EH, Detitta GT. Expert Opin Drug Discov. 2011 May;6(5):465-80.
Introduction: X-ray crystallography provides the majority of our structural biological knowledge at a molecular level and, in terms of pharmaceutical design, is a valuable tool to accelerate discovery. It is the premier technique in the field, but its usefulness is significantly limited by the need to grow well-diffracting crystals. It is for this reason that high-throughput crystallization has become a key technology that has matured over the past 10 years through the field of structural genomics.
Areas covered: The authors describe their experiences in high-throughput crystallization screening in the context of structural genomics and the general biomedical community. They focus on the lessons learnt from the operation of a high-throughput crystallization-screening laboratory, which to date has screened over 12,500 biological macromolecules. They also describe the approaches taken to maximize the success while minimizing the effort. Through this, the authors hope that the reader will gain an insight into the efficient design of a laboratory and protocols to accomplish high-throughput crystallization on a single-, multiuser laboratory or industrial scale. Expert opinion: High-throughput crystallization screening is readily available but, despite the power of the crystallographic technique, getting crystals is still not a solved problem. High-throughput approaches can help when used skillfully; however, they still require human input in the detailed analysis and interpretation of results to be more successful.
Small angle X-ray scattering as a complementary tool for high-throughput structural studies. Grant TD, Luft JR, Wolfley JR, Tsuruta H, Martel A, Montelione GT, Snell EH. Biopolymers. 2011 Aug;95(8):517-30.
Demonstrating the complementarity of SAXS to crystallography and the additional information it can provide.
Structural crystallography and nuclear magnetic resonance (NMR) spectroscopy are the predominant techniques for understanding the biological world on a molecular level. Crystallography is constrained by the ability to form a crystal that diffracts well and NMR is constrained to smaller proteins. Although powerful techniques, they leave many soluble, purified structurally uncharacterized protein samples. Small angle X-ray scattering (SAXS) is a solution technique that provides data on the size and multiple conformations of a sample, and can be used to reconstruct a low-resolution molecular envelope of a macromolecule. In this study, SAXS has been used in a high-throughput manner on a subset of 28 proteins, where structural information is available from crystallographic and/or NMR techniques. These crystallographic and NMR structures were used to validate the accuracy of molecular envelopes reconstructed from SAXS data on a statistical level, to compare and highlight complementary structural information that SAXS provides, and to leverage biological information derived by crystallographers and spectroscopists from their structures. All the ab initio molecular envelopes calculated from the SAXS data agree well with the available structural information. SAXS is a powerful albeit low-resolution technique that can provide additional structural information in a high-throughput and complementary manner to improve the functionalinterpretation of high-resolution structures.
Crystal cookery – using high-throughput technologies and the grocery store as a teaching tool. Luft JR, Furlani NM, Nemoyer RE, Penna EJ, Wolfley JR, Snell ME, Potter SA, Snell EH. J Appl Crystallogr. 2010 Oct 1;43(Pt 5):1189-1207.
An educational paper involving teaching the scientific method through crystallization.
Crystallography is a multidisciplinary field that links divergent areas of mathematics, science and engineering to provide knowledge of life on an
atomic scale. Crystal growth, a key component of the field, is an ideal vehicle for education. Crystallization has been used with a ‘grocery store chemistry’ approach and linked to high-throughput remote-access screening technologies. This approach provides an educational opportunity that can effectively teach the scientific method, readily accommodate different levels of educational experience, and reach any student with access to a grocery store, a post office and the internet. This paper describes the formation of the program through the students who helped develop and prototype the procedures. A summary is presented of the analysis and preliminary results and a description given of how the program could be linked with other aspects of crystallography. This approach has the potential to bridge the gap between students in remote locations and with limited funding, and access to scientific resources, providing students with an international-level research experience.
Macromolecular Crystallization and Crystal Perfection. Chayen, NE, Helliwell, JR, and Snell EH. IUCr Monographs in Crystallography Number 24, Oxford University Press (2010).
The crystallization of proteins and nucleic acids and/or their complexes has become more highly automated but is still often a trial and error based approach. In parallel, a number of X-ray diffraction based techniques have been developed which explain the physical reasons limiting the resulting crystallographic data and thus show how that data may be improved. Crystal growth is also pivotal in neutron crystallography, which establishes the hydrogen and hydration aspects. Thus this book is aimed at addressing the science behind obtaining the best and most complete structural data possible for biological macromolecules, so that the detailed structural biology and chemistry of these important molecules emerge. Crystal imperfections such as twinning and lattice disorders, as well as multiple crystal situations, and their possible remedies, are also described. The small crystal frontier in micro-crystal crystallography, crystallites in powders and finally down to the proposed single molecule structure determination of X-ray lasers are covered. Overall this interdisciplinary book will interest crystal growers, X-ray and neutron physicists and the biological crystallographers, including graduate students.
Sliding clamp-DNA interactions are required for viability and contribute to DNA polymerase management in Escherichia coli. Heltzel JM, Scouten Ponticelli SK, Sanders LH, Duzen JM, Cody V, Pace J, Snell EH, Sutton MD. J Mol Biol. 2009 Mar 20;387(1):74-91.
Sliding clamp proteins topologically encircle DNA and play vital roles in
coordinating the actions of various DNA replication, repair, and damage
tolerance proteins. At least three distinct surfaces of the Escherichia coli β clamp interact physically with the DNA that it topologically encircles. We utilized mutant β clamp proteins bearing G66E and G174A substitutions (β159), affecting the single-stranded DNA-binding region, or poly-Ala substitutions in place of residues 148-HQDVR-152 (β148–152), affecting the double-stranded DNA binding region, to determine the biological relevance of clamp–DNA interactions. As part of this work, we solved the X-ray crystal structure of β148–152, which verified that the poly-Ala substitutions failed to significantly alter the tertiary structure of the clamp. Based on functional assays, both β159 and β148–152 were impaired for loading and retention on a linear primed DNA in vitro. In the case of β148–152, this defect was not due to altered interactions with the DnaX clamp loader, but rather was the result of impaired β148–152–DNA interactions. Once loaded, β148–152 was proficient for DNA polymerase III (Pol III) replication in vitro. In contrast, β148–152 was severely impaired for Pol II and Pol IV replication and was similarly impaired for direct physical interactions with these Pols. Despite its ability to support Pol III replication in vitro, β148–152 was unable to support viability of E. coli. Nevertheless, physiological levels of β148–152 expressed from a plasmid efficiently complemented the temperaturesensitive growth phenotype of a strain expressing β159 (dnaN159), provided that Pol II and Pol IV were inactivated. Although this strain was impaired for Pol V-dependent mutagenesis, inactivation of Pol II and Pol IV restored the Pol V mutator phenotype. Taken together, these results support a model in which a sophisticated combination of competitive clamp–DNA, clamp–partner, and partner–DNA interactions serve to manage the actions of the different E. coli Pols in vivo
Establishing a training set through the visual analysis of crystallization trials. Part II: crystal examples. Snell EH, Lauricella AM, Potter SA, Luft JR, Gulde SM, Collins RJ, Franks G, Malkowski MG, Cumbaa C, Jurisica I, DeTitta GT. Acta Crystallogr D Biol Crystallogr. 2008 Nov;64(Pt 11):1131-7.
Part of an effort to build a robust training set for image analysis and machine learning – crystal examples.
In the automated image analysis of crystallization experiments, representative examples of outcomes can be obtained rapidly. However, while the outcomes appear to be diverse, the number of crystalline outcomes can be small. To complement a training set from the visual observation of 147 456 crystallization outcomes, a set of crystal images was produced from 106 and 163 macromolecules under study for the North East Structural Genomics Consortium (NESG) and Structural Genomics of Pathogenic Protozoa (SGPP) groups, respectively. These crystal images have been combined with the initial training set. A description of the crystal-enriched data set and a preliminary analysis of outcomes from the data are described.
Establishing a training set through the visual analysis of crystallization trials. Part I: approximately 150,000 images. Snell EH, Luft JR, Potter SA, Lauricella AM, Gulde SM, Malkowski MG, Koszelak-Rosenblum M, Said MI, Smith JL, Veatch CK, Collins RJ, Franks G, Thayer M, Cumbaa C, Jurisica I, Detitta GT. Acta Crystallogr D Biol Crystallogr. 2008 Nov;64(Pt 11):1123-30.
Part of an effort to build a robust training set for image analysis and machine learning – typical outcomes.
Structural crystallography aims to provide a three-dimensional representation of macromolecules. Many parts of the multistep process to produce the three-dimensional structural model have been automated, especially through various structural genomics projects. A key step is the production of crystals for diffraction. The target macromolecule is combined with a large and chemically diverse set of cocktails with some leading ideally, but infrequently, to crystallization. A variety of outcomes will be observed during these screening experiments that typically require human interpretation for classification. Human interpretation is neither scalable nor objective, highlighting the need to develop an automatic computer based image classification. As a first step towards automated image classification, 147 456 images representing crystallization experiments from 96 different macromolecular samples were manually classified. Each image was classified by three experts into seven predefined categories or their combinations. The resulting data where all three observers are in agreement provides one component of a truth set for the development and rigorous testing of automated imageclassification systems and provides information about the chemical cocktails used for crystallization. In this paper, the details of this study are presented.
Glycerol concentrations required for the successful vitrification of cocktail conditions in a high-throughput crystallization screen. Kempkes R, Stofko E, Lam K, Snell EH. Acta Crystallogr D Biol Crystallogr. 2008 Mar;64(Pt 3):287-301.
The Hauptman–Woodward Medical Research Institute runs a high-throughput crystallization screening service in which macromolecules are screened against 1536 potential crystallization cocktails. Typically, multiple crystallization leads are identified. With a limited amount of sample, the question becomes ‘How many leads can be optimized and which leads are most likely to produce X-ray diffraction data?’. In order to prioritize the hits for optimization, the amount of glycerol required to successfully cryocool each cocktail has been determined for the cocktails used in the high-throughput screen. Those hit conditions that require the minimum amount of cryoprotectant for successful vitrification will be closer in chemical make-up to the mother liquor. Hence, if the physical properties of the crystals are similar, one could logically prioritize leads that are more likely to produce diffraction based upon the chemical similarity of the native to the cryopreserved mother liquor.
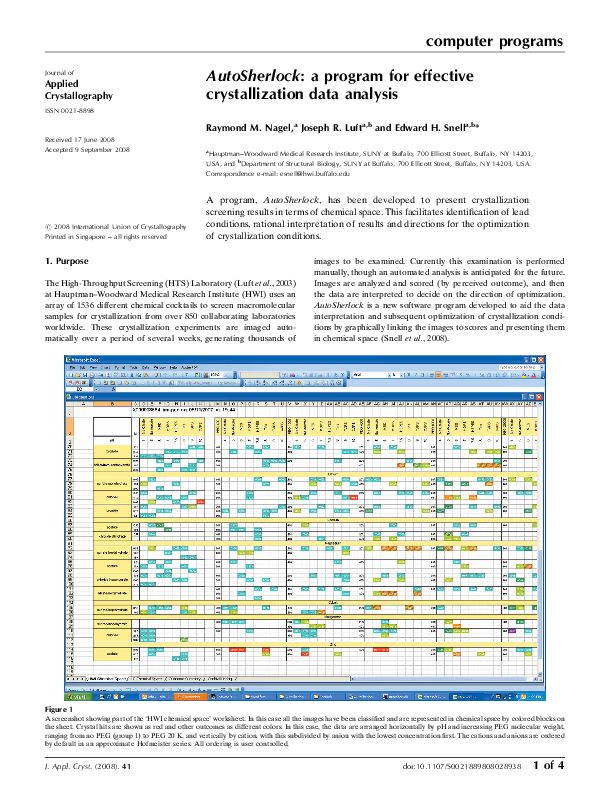
The application and use of chemical space mapping to interpret crystallization screening results. Snell EH, Nagel RM, Wojtaszcyk A, O’Neill H, Wolfley JL, Luft JR. Acta Crystallogr D Biol Crystallogr. 2008 Dec;64(Pt 12):1240-9.
Macromolecular crystallization screening is an empirical process. It often begins by setting up experiments with a number of chemically diverse cocktails designed to sample chemical space known to promote crystallization. Where a potential crystal is seen a refined screen is set up, optimizing around that condition. By using an incomplete factorial sampling of chemical space to formulate the cocktails and presenting the results graphically, it is possible to readily identify trends relevant to crystallization, coarsely sample the phase diagram and help guide the optimization process. In this paper, chemical space mapping is applied to both single macromolecules and to a diverse set of macromolecules in order to illustrate how visual information is more readily understood and assimilated than the same information presented textually
AutoSherlock: a program for effective crystallization data analysis. Nagel RM, Luft JR, Snell EH. J Appl Crystallogr. 2008 Dec 1;41(Pt 6):1173-1176.
A program, AutoSherlock, has been developed to present crystallization screening results in terms of chemical space. This facilitates identification of lead
conditions, rational interpretation of results and directions for the optimization of crystallization conditions.
Digital X-ray camera for quality evaluation three-dimensional topographic reconstruction of single crystals of biological macromolecules. Borgstahl, G., Lovelace, J., Snell, E.H., Bellamy, H. 7,466,798, Issue Dec. 16th 2008.
The present invention provides a digital topography imaging system for determining the crystalline structure of a biological macromolecule, wherein the system employs a charge coupled device (CCD) camera with antiblooming circuitry to directly convert X-ray signals without the use of phosphor and measure reflection profiles from the X-ray emitting source after X-ray are passed through a sample. Methods for using said system are also provided.
Efficient optimization of crystallization conditions by manipulation of drop volume ratio and temperature. Luft JR, Wolfley JR, Said MI, Nagel RM, Lauricella AM, Smith JL, Thayer MH, Veatch CK, Snell EH, Malkowski MG, Detitta GT. Protein Sci. 2007 Apr;16(4):715-22.
An efficient optimization method for the crystallization of biological macromolecules has been developed and tested. This builds on a successful high-throughput technique for the determination of initial crystallization conditions. The optimization method takes an initial condition identified through screening and then varies the concentration of the macromolecule, precipitant, and the growth temperature in a systematic manner. The amount of sample and number of steps is minimized and no biochemical reformulation is required. In the current application a robotic liquid handling system enables highthroughput use, but the technique can easily be adapted in a nonautomated setting. This method has been applied successfully for the rapid optimization of crystallization conditions in nine representative cases.
Changes to crystals of Escherichia coli beta-galactosidase during room-temperature/low-temperature cycling and their relation to cryo-annealing. Juers DH, Lovelace J, Bellamy HD, Snell EH, Matthews BW, Borgstahl GE. Acta Crystallogr D Biol Crystallogr. 2007 Nov;63(Pt 11):1139-53.
Flash-cooling of macromolecular crystals often compromises diffraction quality by increasing the mosaicity. In some cases, cycling the crystal between low temperature (LT) and room temperature (RT) can reverse this increase in mosaicity. Previous studies of RT/LT cycling have focused on the quality of the crystal as it was repeatedly returned to the LT state. Here, crystal quality is explored not only at LT but also when the crystal is returned to RT. The domain model is used to extract information about crystal order from reflection profiles measured from crystals of Escherichia coli β-galactosidase at both temperatures. Despite optimization of the cryocooling protocol, the mosaicity increases by about sixfold with cooling and is anisotropic at both temperatures. The mosaicity increase is the consequence of a decrease in domain volume, an increase in the variation of domain cell dimensions and an increase in the angular spread between domains. Upon rewarming, the mosaicity recovers substantially, including the somewhat surprising recovery of domain volume, but incompletely. Over multiple RT/LT cycles disorder in both states increases, which appears to mainly arise from radiation damage, although a contribution from cool–thaw processes cannot be ruled out. The analysis further suggests that LT disorder is governed by variability inherent in the cooling process combined with the overall history of the crystal. In contrast, RT disorder appears to be governed principally by the overall history of the crystal. This suggests that with these particular crystals under the experimental conditions used, particularly at high-intensity synchrotron X-ray sources, RT/LT cycling annealing protocols should involve few cycles so as to limit the hysteresis in both temperature states while taking advantage of the inherent variability in the cooling process that may result in improved crystal order at LT.
Non-invasive measurement of X-ray beam heating on a surrogate crystal sample. Snell EH, Bellamy HD, Rosenbaum G, van der Woerd MJ. J Synchrotron Radiat. 2007 Jan;14(Pt 1):109-15.
The first non-invasive measurements to evaluate protein crystal heating in a third-generation synchrotron beam.
Cryocooling is a technique routinely used to mitigate the effects of secondary radiation damage on macromolecules during X-ray data collection. Energy from the X-ray beam absorbed by the sample raises the temperature of the sample. How large is the temperature increase and does this reduce the effectiveness of cryocooling? Sample heating by the X-ray beam has been measured noninvasively for the first time by means of thermal imaging. Specifically, the temperature rise of 1 mm and 2 mm glass spheres (sample surrogates) exposed to an intense synchrotron X-ray beam and cooled in a laminar flow of nitrogen gas is experimentally measured. For the typical sample sizes, photon energies, fluxes, flux densities and exposure times used for macromolecular crystallographic data collection at third-generation synchrotron radiation sources and with the sample accurately centered in the cryostream, the heating by the X-ray beam is only a few degrees. This is not sufficient to raise the sample above the amorphous-ice/crystalline-ice transition temperature and, if the cryostream cools the sample to 100 K, not even enough to significantly enhance radiation damage from secondary effects.
Structure of the full-length human RPA14/32 complex gives insights into the mechanism of DNA binding and complex formation. Deng X, Habel JE, Kabaleeswaran V, Snell EH, Wold MS, Borgstahl GE. J Mol Biol. 2007 Dec 7;374(4):865-76.
Replication protein A (RPA) is the ubiquitous, eukaryotic single-stranded
DNA (ssDNA) binding protein and is essential for DNA replication, recombination, and repair. Here, crystal structures of the soluble RPA heterodimer, composed of the RPA14 and RPA32 subunits, have been determined for the full-length protein in multiple crystal forms. In all crystals, the electron density for the N-terminal (residues 1–42) and C-terminal (residues 175–270) regions of RPA32 is weak and of poor quality indicating that these regions are disordered and/or assume multiple positions in the crystals. Hence, the RPA32 N terminus, that is hyperphosphorylated in a cell-cycle-dependent manner and in response to DNA damaging agents, appears to be inherently disordered in the unphosphorylated state. The C-terminal, winged helix-loop-helix, protein–protein interaction domain adopts several conformations perhaps to facilitate its interaction with various proteins. Although the ordered regions of RPA14/32 resemble the previously solved protease-resistant core crystal structure, the quaternary structures between the heterodimers are quite different. Thus, the four-helix bundle quaternary assembly noted in the original core structure is unlikely to be related to the quaternary structure of the intact heterotrimer. An organic ligand binding site between subunits RPA14 and RPA32 was identified to bind dioxane. Comparison of the ssDNA binding surfaces of RPA70 with RPA14/32 showed that the lower affinity of RPA14/32 can be attributed to a shallower binding crevice with reduced positive electrostatic charge.
Optimizing crystal volume for neutron diffraction: D-xylose isomerase. Snell EH, van der Woerd MJ, Damon M, Judge RA, Myles DA, Meilleur F. Eur Biophys J. 2006 Sep;35(7):621-32.
Neutron diffraction is uniquely sensitive to hydrogen positions and protonation state. In that context structural information from neutron data is complementary to that provided through X-ray diffraction. However, there are practical obstacles to overcome in fully exploiting the potential of neutron diffraction, i.e. low flux and weak scattering. Several approaches are available to overcome these obstacles and we have investigated the simplest: increasing the diffracting volume of the crystals. Volume is a quantifiable metric that is well suited for experimental design and optimization techniques. By using response surface methods we have optimized the xylose isomerase crystal volume, enabling neutron diffraction while we determined the crystallization parameters with a minimum of experiments. Our results suggest a systematic means of enabling neutron diffraction studies for a larger number of samples that require information on hydrogen position and/or protonation state.
A quasi-Laue neutron crystallographic study of D-xylose isomerase. Meilleur F, Snell EH, van der Woerd MJ, Judge RA, Myles DA. Eur Biophys J. 2006 Sep;35(7):601-9.
The location of hydrogen atoms in enzyme structures can bring critical understanding of catalytic mechanism. However, whilst it is often difficult to determine the position of hydrogen atoms using X-ray crystallography even with subatomic ( < 1.0 Å ) resolution data available, neutron crystallography provides an experimental tool to directly localize hydrogen/deuterium atoms in biological macromolecules at resolution of 1.5–2.0 Å. D-Xylose isomerase (D-xylose ketol-isomerase, EC 5.3.1.5) is a 43 kDa enzyme that catalyses the first reaction in the catabolism of D-xylose. Linearization and isomerization of D-xylose at the active site of D-xylose isomerase rely upon a complex hydrogen transfer. Neutron quasi-Laue data at 2.2 Å resolution were collected at room temperature on a partially deuterated Streptomyces rubiginosus D-xylose isomerase crystal using the LADI instrument at ILL with the objective to provide insight into the enzymatic mechanism. The neutron structure shows unambiguously that residue His 53 is doubly protonated at the active site of the enzyme. This suggests that the reaction proceeds through an acid catalyzed opening of the sugar ring, which is in accord with the mechanism suggested by Fenn et al. (Biochemistry 43(21): 6464–6474, 2004). This is the first report of direct observation of double protonation of His 53 and the first validation of the ring opening mechanism at the active site of D-xylose isomerase.
Finding a cold needle in a warm haystack: Infrared imaging applied to locating cryocooled crystals. Snell, EH, van der Woerd, MJ, Miller, MD and Deacon, AM. Journal of Applied Crystallography, 38, 69-77 (2005).
The use of infrared imaging to locate crystals mounted in cryoloops and
cryopreserved in a nitrogen gas stream at 100 K is demonstrated. In the home laboratory, crystals are clearly seen in the infrared images with light transmitting through the sample while irradiating the crystal from behind, and with illumination from a direction perpendicular to the direction of view. The crystals transmit and reflect infrared radiation at different levels to the surrounding mother liquor and loop. Because of differences in contrast between crystals and their surrounding mother liquor, it is possible to identify the crystal position. At the synchrotron, with robotically mounted crystals, the small depth of field of the lens required the recording of multiple images at different focal points. Image processing techniques were then used to construct a clear image of the crystal. The resulting infrared images and intensity profiles show that infrared imaging can be a powerful complement to visual imaging in locating crystals in cryocooled loops.
Crystallization in Microgravity. Snell, EH, and Helliwell, JR. Macromolecular Reports on Progress in Physics, 68, 799-853 (2005).
Density difference fluid flows and sedimentation of growing crystals are greatly reduced when crystallization takes place in a reduced gravity environment. In the case of macromolecular crystallography a crystal of a biological macromolecule is used for diffraction experiments (x-ray or neutron) so as to determine the three-dimensional structure of the macromolecule. The better the internal order of the crystal then the greater the molecular structure detail that can be extracted. It is this structural information that enables an understanding of how the
molecule functions. This knowledge is changing the biological and chemical sciences, with major potential in understanding disease pathologies.
In this review, we examine the use of microgravity as an environment to grow macromolecular crystals. We describe the crystallization procedures used on the ground, how the resulting crystals are studied and the knowledge obtained from those crystals. We address the features desired in an ordered crystal and the techniques used to evaluate those features in detail. We then introduce the microgravity environment, the techniques to access that environment and the theory and evidence behind the use of microgravity for crystallization experiments. We describe how ground-based laboratory techniques have been adapted to
microgravity flights and look at some of the methods used to analyse the resulting data. Several case studies illustrate the physical crystal quality improvements and the macromolecular structural advances. Finally, limitations and alternatives to microgravity and future directions for this research are covered.
Macromolecular structural crystallography in general is a remarkable field where physics, biology, chemistry and mathematics meet to enable insight to the fundamentals of life. As the reader will see, there is a great deal of physics involved when the microgravity environment is applied to crystallization, some of it known, and undoubtedly much yet to discover.
Extracting trends from two decades of microgravity macromolecular crystallization history. Judge RA, Snell EH, van der Woerd MJ. Acta Crystallogr D Biol Crystallogr. 2005 Jun;61(Pt 6):763-71.
Since the 1980s hundreds of macromolecular crystal growth experiments have been performed in the reduced acceleration environment of an orbiting spacecraft. Significant enhancements in structural knowledge have resulted from X-ray diffraction of the crystals grown. Similarly, many samples have shown no improvement or degradation in comparison to those grown on the ground. A complex series of interrelated factors affect these experiments and by building a comprehensive archive of the results it was aimed to identify factors that result in success and those that result in failure. Specifically, it was found that dedicated microgravity missions increase the chance of success when compared with those where crystallization took place as a parasitic aspect of the mission. It was also found that the chance of success could not be predicted based on any discernible property of the macromolecule available to us.
Imaging modulated reflections from a semi-crystalline state of profiling:actin. Lovelace, JJ, Narayan, K, Chik, JK, Bellamy, HD, Snell, EH, Lindberg, U, Schutt, CE and Borgstahl, GEO.J. Applied Crystallography 37, 327-330 (2004).
Modulated protein crystals remain terra incognita for most crystallographers. While small-molecule crystallographers have successfully wrestled with and conquered this type of structure determination, to date no modulated macromolecular structures have been reported. Proflin:β-actin in a modulated semi-crystalline state presents a challenge of suficient biological signi®cance to motivate the development of methods for the accurate collection of data on the complex diffraction pattern and, ultimately, the solution of its structure. In the present work, fine-sliced data collection was used to resolve the closely spaced satellite reflections from these polymorphic crystals. Image-processing methods were used to visualize these data for comparison with the original precession data. These preliminary data demonstrate the feasibility of using fine -slicing to collect accurately the intensities and positions of the main and satellite reflections from these modulated protein crystals.
First results of digital topography applied to macromolecular crystals. Lovelace, JJ, Soares, A, Bellamy, HD, Sweet, RM, Snell, EH and Borgstahl, GEO. J. Applied Crystallography 37, 481-485 (2004).
An inexpensive digital CCD camera was used to record X-ray topographs
directly from large imperfect crystals of cubic insulin. The topographs recorded were not as detailed as those which can be measured with ®lm or emulsion plates, but do show great promise. Six rflections were recorded using a set of finely spaced stills encompassing the rocking curve of each reflection. A complete topographic reflection pro®le could be digitally imaged in minutes. Interesting and complex internal structure was observed by this technique. The CCD chip used in the camera has anti-blooming circuitry and produced good data quality, even when pixels became overloaded.
Physical and structural studies on the cryocooling of insulin crystals. Vahedi-Faridi A, Lovelace J, Bellamy HD, Snell EH, Borgstahl GE. Acta Crystallogr D Biol Crystallogr. 2003 Dec;59(Pt 12):2169-82.
Reflection profiles were analyzed from microgravity-grown (mg) and earth-grown insulin crystals to measure mosaicity (η) and to reveal mosaic domain structure and composition. The effects of cryocooling on single-domain and multi-domain crystals were compared. The effects of cryocooling on insulin structure were also re-examined. Microgravity crystals were of larger volume, were more homogeneous and were of higher quality than earth crystals. Several mg crystals contained a single mosaic domain which encompassed all or nearly all of the crystal with an η avg of 0.005°. The earth crystals varied in quality and all contained multiple domains with an η avg of 0.031°. Cryocooling caused a 43-fold increase in η for mg crystals (η avg = 0.217°) and an eightfold increase for earth crystals (η avg = 0.246°). These results indicate that very well ordered crystals are not completely protected from the stresses associated with cryocooling, especially when structural perturbations occur. However, there were differences in the reflection profiles. For multi-mosaic domain crystals, each domain individually broadened and separated from the other domains upon cryocooling. Cryocooling did not cause an increase in the number of domains. A crystal composed of a single domain retained this domain structure and the reflection profiles simply broadened. Therefore, an improved signal-to-noise ratio for each reflection was measured from cryocooled single-domain crystals relative to cryocooled multi-domain crystals. The improved signal from mg crystals, along with the increase in crystal size, facilitated the measurement of the weaker high-resolution reflections. The observed broadening of reflection pro®les indicates increased variation in unit-cell parameters, which may be linked to cryocooling-associated structural changes and disorder.
Macromolecular crystal quality. Snell EH, Bellamy HD, Borgstahl GE. Methods Enzymol. 2003;368:268-88.
The internal order of a crystal can be characterized by a correlation
length, i.e., the distance over which all the atoms in unit cells are ‘‘accurately’’ related by the crystal-symmetry operators. An atom will contribute coherently to the intensity of a reflection only if its disorder relative to symmetry-related atoms is small compared to the resolution of the reflection. For a constant average random disorder in atomic position between adjacent unit cells, the disorder between any two unit cells will increase as the square root of the distance between them. Therefore as resolution increases the effective correlation length decreases, and the number of unit cells contributing coherently to the diffraction decreases.
Random disorder is a major contributor to the reduction in diffracted
intensity with increasing resolution. Disorder can be described as long range or short range. In general long-range disorder in the crystal gives rise to localized effects in reciprocal space and vice versa.For example, crystal mosaicity, which is a large-scale property in real space, causes the localized effect of broadened spots in reciprocal space. Random disorder between adjacent unit cells, a short-scale property in real space, is seen as a global, resolution-dependent reduction in diffracted intensity in reciprocal space. Since macromolecular crystals are, by the standard of small molecule crystals, not very good crystals, they offer a fruitful field for the study of disorder and how to minimize that disorder.
The development and application of a method to quantify the quality of cryoprotectant conditions using standard area detector X-ray images. McFerrin, M and Snell, EH. J. Appl. Cryst. 35, 538-545 (2002).
An X-ray based method for determining initial cryoprotectant concentrations necessary to protect solutions from crystalline ice formation on flash cooling was developed. X-ray images from a charge-coupled device (CCD) area detector were integrated as powder patterns and quantified by determining the standard deviation of the slope of the normalized intensity curve in the resolution range where ice rings are known to occur. The method was tested by determining the concentrations of glycerol, PEG400, ethylene glycol and 1,2-propanediol
necessary to form an amorphous glass at 100 K with each of the 98 crystallization solutions of Crystal Screens I and II (Hampton Research, Laguna Hills, California, USA). For conditions that required glycerol concentrations of 35% or above, cryoprotectant conditions using (2R,3R)-(–)-2,3-butanediol were determined. The method proved to be remarkably reliable. The results build on previous work [Garman & Mitchell (1996). J. Appl. Cryst. 29, 584-587] and extend the number of suitable starting conditions to alternative cryoprotectants.
Seeing the heat — preliminary studies of cryocrystallography using infrared imaging. Snell EH, Judge RA, Larson M, van der Woerd MJ. J Synchrotron Radiat. 2002 Nov 1;9(Pt 6):361-7.
The first imaging of the cryocoolin process in a protein crystal.
As preparation for an extensive study that aims to image the cryocooling process of macromolecular crystals, the ability to thermally image solid objects and liquids at temperatures far below 273 K is demonstrated. In the case of a large lysozyme crystal (1.0 x 0.7 x 0.2 mm), qualitative measurements show the cooling process to take about 0.6 s with the cooling taking place in a wave starting from the face of the crystal nearest to the origin of the cryostream and ending at the point furthest away from the origin. Annealing of this lysozyme crystal, cooled under good cryoprotectant conditions, shows that cold striations form perpendicular to the cooling stream. These striations become more pronounced after successive annealing. Cryocooling of a non-cryoprotected crystal of glucose isomerase displayed an `S-shaped’ cold front wave traveling across the sample. These preliminary results are qualitative but show the power of infrared imaging as a new tool for fundamental and practical cryocrystallography studies.
Thaumatin crystallization aboard the International Space Station using liquid-liquid diffusion in the Enhanced Gaseous Nitrogen Dewar (EGN). Barnes CL, Snell EH, Kundrot CE.Acta Crystallogr D Biol Crystallogr. 2002 May;58(Pt 5):751-60.
This paper reports results from the first biological crystal-growth experiment on the International Space Station (ISS). Crystals of thaumatin were grown using liquid-liquid diffusion in Tygon tubing transported in the Enhanced Gaseous Nitrogen Dewar (EGN). Different volume ratios and concentrations of protein and precipitant were used to test different adaptations of the vapor-diffusion crystallization recipe to the liquid-liquid diffusion method. The EGN warmed up from 77 to 273 K in about 4 d, about the same time it took to warm from 273 to 293 K. The temperature within the EGN was 293-297 K for the majority of the experiment. Air gaps that blocked liquid-liquid diffusion formed in the tubes. Nonetheless, crystals were grown. Synchrotron diffraction data collected from the best space-grown crystal extended to 1.28 Å, comparable to previous studies of space-grown thaumatin crystals. The resolution of the best ground-control crystal was only 1.47 Å. It is not clear if the difference in diffraction limit arises from factors other than crystal size. Improvements in temperature control and the elimination of air gaps are needed, but the results show that the EGN on the ISS can be used to produce space-grown crystals that diffract to high resolution.
Free-falling crystals: Biological macromolecular crystal growth studies in low earth orbit. Judge, RA., Snell, EH. and Pusey, ML. Dev. Chem. Eng. Mineral Process. 10(5/6) 479-488 (2002).
Spacecraft orbiting the earth experience a reduced acceleration environment due to being in a state of continuous free-fall. This state colloquially termed microgravity, has produced improved X-ray difiaction quality crystals of biological macromolecules. Improvements in X-ray difiaction resolution (or detail) or signal to noise, provide greater detail in the three-dimensional molecular structure providing information about the molecule, how it works, how to improve its fitnction or how to impede it. Greater molecular detail obtained by crystallization in microgravity, has important implications for structural biology. In this paper we examine the theories behind macromolecule crystal quality improvement in microgravity using results obtained frm studies with the model protein, chicken egg white lysozyme.
Fluid Flows and Macromolecular Growth in Microgravity,Helliwell, JR, Snell, EH, Chayen, NE, Judge, RA, Boggon, TJ and Pusey, ML. Published in Physics of Fluids in Microgravity, ch 14, pages 489-514. Taylor and Francis. Editor, R. Monti (2002).
Method for measurement of physical characteristics of crystals. Borgstahl, G., Lovelace, J., Snell, E.H. 6,498,829, Issue date Dec. 24th 2002.
A method and apparatus to simultaneously measure the diffractive resolution and mosaic spread of macromolecular crystals, are described. The method includes minimizing contributions of an X-ray beam to any reflection angular widths in the crystal, repidly measuring multi reflection profiles in the crystal over a wide resolution range, evaluating and deconvoluting the Lorentz effect and beam contributions, and determining the direction in which the crystal is most perfect.
A test of macromolecular crystallization in microgravity: large well ordered insulin crystals. Borgstahl GE, Vahedi-Faridi A, Lovelace J, Bellamy HD, Snell EH. Acta Crystallogr D Biol Crystallogr. 2001 Aug;57(Pt 8):1204-7.
Stastical confirmation of the increase in physical quality of the crystal from microgravity growth.
Crystals of insulin grown in microgravity on Space Shuttle Mission STS-95 were extremely well ordered and unusually large (many >2 mm). The physical characteristics of six microgravity and six earth-grown crystals were examined by X-ray analysis employing superfine phi slicing and unfocused synchrotron radiation. This experimental setup allowed hundreds of reflections to be precisely examined from each crystal in a short period of time. The microgravity crystals were on average 34 times larger, had sevenfold lower mosaicity, had 54-fold higher reflection peak heights and diffracted to significantly higher resolution than their earth-grown counterparts. A single mosaic domain model could account for the observed reflection profiles in microgravity crystals, whereas data from earth crystals required a model with multiple mosaic domains. This statistically significant and unbiased characterization indicates that the microgravity environment was useful for the improvement of crystal growth and the resultant diffraction quality in insulin crystals and may be similarly useful for macromolecular crystals in general.
Investigating the effect of impurities on macromolecule crystal growth in microgravity. Snell, EH, Judge, RA, Crawford, L, Forsythe, EL, Pusey, ML, Sportiello, M, Todd, P, Bellamy, H, Lovelace, J, Cassanto, Borgstahl, GEO. Crystal Growth and Design, 1, 2, 151-158 (2001).
Chicken egg-white lysozyme (CEWL) crystals were grown in microgravity and on the ground in the presence of various amounts of a naturally occurring lysozyme dimer impurity. No significant favorable differences in impurity incorporation between microgravity and ground crystal samples were observed. At low impurity concentration the microgravity crystals preferentially incorporated the dimer. The presence of the dimer in the crystallization solutions in microgravity reduced crystal size, increased mosaicity, and reduced the signal-to-noise ratio of the X-ray data. Microgravity samples proved more sensitive to impurity. Accurate indexing of the reflections proved critical to the X-ray analysis. The largest crystals with the best X-ray diffraction properties were grown from pure solution in microgravity.
Microgravity and Macromolecular Crystallography. Kundrot, CE, Judge, RA, Pusey, ML, & Snell, EH. Crystal Growth and Design. Crystal Growth and Design, 1, 87-99 (2001).
Macromolecular crystal growth has been seen as an ideal experiment to make use of the reduced acceleration environment provided by an orbiting spacecraft. The experiments are small, simply operated and have a high potential scientific and economic impact. In this review we examine the theoretical reasons why microgravity should be a beneficial environment for crystal growth and survey the history of experiments on the Space Shuttle Orbiter, on unmanned spacecraft, and on the Mir space station. Finally we outline the direction for optimizing the future use of orbiting platforms.
The high-mosaicity illusion: revealing the true physical characteristics of macromolecular crystals. Bellamy HD, Snell EH, Lovelace J, Pokross M, Borgstahl GE. Acta Crystallogr D Biol Crystallogr. (2000) Aug;56(Pt 8):986-95.
Typical measurements of macromolecular crystal mosaicity are dominated by the characteristics of the X-ray beam and as a result the mosaicity value given during data processing can be an artifact of the instrumentation rather than the sample. For physical characterization of crystals, an experimental system and software have been developed to simultaneously measure the diffraction resolution and mosaic spread of macromolecular crystals. The contributions of the X-ray beam to the reflection angular widths were minimized by using a highly parallel, highly monochromatic synchrotron source. Hundreds of reflection profiles over a wide resolution range were rapidly measured using a charge-coupled device (CCD) area detector in combination with superfine phi-slicing data collection. The Lorentz effect and beam contributions were evaluated and deconvoluted from the recorded data. Data collection and processing is described. From 1 degrees of superfine phi-slice data collected on a crystal of manganese superoxide dismutase, the mosaicities of 260 reflections were measured. The average mosaicity was 0.0101 degrees (s.d. 0.0035 degrees ) measured as the full-width at half-maximum (FWHM) and ranged from 0.0011 to 0. 0188 degrees. Each reflection profile was individually fitted with two Gaussian profiles, with the first Gaussian contributing 55% (s.d. 9%) and the second contributing 35% (s.d. 9%) of the reflection. On average, the deconvoluted width of the first Gaussian was 0.0054 degrees (s.d. 0.0015 degrees ) and the second was 0.0061 degrees (s. d. 0.0023 degrees ). The mosaicity of the crystal was anisotropic, with FWHM values of 0.0068, 0.0140 and 0.0046 degrees along the a, b and c axes, respectively. The anisotropic mosaicity analysis indicates that the crystal is most perfect in the direction that corresponds to the favored growth direction of the crystal.
BEAM-ish: A graphical user interface for the physical characterization of macromolecular crystals. Lovelace, J, Snell, EH, Pokross, M, Arvai, A, Nielsen, C, Nguyen, X, Bellamy, H and Borgstahl, GEO. J. Applied Crystallography, 33, 1187-1188 (2000).
Synchrotron X-ray reciprocal-space mapping, topography and diffraction resolution studies of macromolecular crystal quality. Boggon TJ, Helliwell JR, Judge RA, Olczak A, Siddons DP, Snell EH, Stojanoff V.Acta Crystallogr D Biol Crystallogr. 2000 Jul;56(Pt 7):868-80.
An example of semiconductor crystal analysis techniques appled to protein crystals and the quality of those crystals.
A comprehensive study of microgravity and ground-grown chicken egg-white lysozyme crystals is presented using synchrotron X-ray reciprocal-space mapping, topography techniques and diffraction resolution. Microgravity crystals displayed reduced intrinsic mosaicities on average, but no differences in terms of strain over their ground-grown counterparts. Topographic analysis revealed that in the microgravity case the majority of the crystal was contributing to the peak of the reflection at the appropriate Bragg angle. In the ground-control case only a small volume of the crystal contributed to the intensity at the diffraction peak. The techniques prove to be highly complementary, with the reciprocal-space mapping providing a quantitative measure of the crystal mosaicity and strain (or variation in lattice spacing) and the topography providing a qualitative overall assessment of the crystal in terms of its X-ray diffraction properties. Structural data collection was also carried out at the synchrotron.
Cryo-trapping the six-coordinate, distorted-octahedral active site of manganese superoxide dismutase. Borgstahl GE, Pokross M, Chehab R, Sekher A, Snell EH. J Mol Biol. 2000 Mar 3;296(4):951-9.
Superoxide dismutase protects organisms from potentially damaging oxygen radicals by catalyzing the disproportionation of superoxide to oxygen and hydrogen peroxide. We report the use of cryogenic temperatures to kinetically capture the sixth ligand bound to the active site of manganese superoxide dismutase (MnSOD). Synchrotron X-ray diffraction data was collected from Escherichia coli MnSOD crystals grown at pH 8.5 and cryocooled to 100 K. Structural refinement to 1.55 A resolution and close inspection of the active site revealed electron density for a sixth ligand that was interpreted to be a hydroxide ligand. The six-coordinate, distorted-octahedral geometry assumed during inhibition by hydroxide is compared to the room temperature, five-coordinate, trigonal bipyramidal active site determined with crystals grown from practically identical conditions. The gateway residues Tyr34, His30 and a tightly bound water molecule are implicated in closing-off the active site and blocking the escape route of the sixth ligand.
Crystallization of chicken egg white lysozyme from assorted sulphate salts. EL Forsythe, EH Snell, CC Malone and ML Pusey. J.Cryst. Growth, 196, 332-343. (1999).
Thought not to be possible from theoretical considerations, lysozyme crystals resulted from sulfate salts.
Chicken egg white lysozyme has been found to crystallize from ammonium, sodium, potassium, rubidium, magnesium, and manganese sulfates at acidic and basic pH, with protein concentrations from 60 to 190 mg/ml. Crystals have also been grown at 4 degree C in the absence of any other added salts using isoionic lysozyme which was titrated to pH 4.6 with dilute sulfuric acid. Four different crystal forms have been obtained, depending upon the temperature, protein concentration, and precipitating salt employed. Crystals grown at 15 degree C were generally tetragonal, with space group P43212. Crystallization at 20 degree C typically resulted in the formation of orthorhombic crystals, space group P212121. The tetragonal reversible reaction orthorhombic transition appeared to be a function of both the temperature and protein concentration, occurring between 15 and 20 degree C and between 100 and 125 mg/ml protein concentration. Crystallization from 1.2 M magnesium sulfate at pH 7.8 gave a trigonal crystal, space group P3121, a = b = 87.4, c = 73.7, gamma = 120 deg, which diffracted to 2.8 A. Crystallization from ammonium sulfate at pH 4.6, generally at lower temperatures, was also found to result in a monoclinic form. space group C2, a = 65.6, b = 95.0, c = 41.2, beta = 119.2 deg. A crystal of approximately 0.2 x 0.2 x 0.5 mm grown from bulk solution diffracted to approximately 3.5 A.
The effect of temperature and solution pH on the nucleation of tetragonal lysozyme crystals. Judge RA, Jacobs RS, Frazier T, Snell EH, Pusey ML. Biophys J. 1999 Sep;77(3):1585-93.
Part of the challenge of macromolecular crystal growth for structure determination is obtaining crystals with a volume suitable for x-ray analysis. In this respect an understanding of the effect of solution conditions on macromolecule nucleation rates is advantageous. This study investigated the effects of supersaturation, temperature, and pH on the nucleation rate of tetragonal lysozyme crystals. Batch crystallization plates were prepared at given solution concentrations and incubated at set temperatures over 1 week. The number of crystals per well with their size and axial ratios were recorded and correlated with solution conditions. Crystal numbers were found to increase with increasing supersaturation and temperature. The most significant variable, however, was pH; crystal numbers changed by two orders of magnitude over the pH range 4.0-5.2. Crystal size also varied with solution conditions, with the largest crystals obtained at pH 5.2. Having optimized the crystallization conditions, we prepared a batch of crystals under the same initial conditions, and 50 of these crystals were analyzed by x-ray diffraction techniques. The results indicate that even under the same crystallization conditions, a marked variation in crystal properties exists.
Crystallization of chicken egg-white lysozyme from ammonium sulfate. Forsythe EL, Snell EH, Pusey ML. Acta Crystallogr D Biol Crystallogr. 1997 Nov 1;53(Pt 6):795-7.
Chicken egg-white lysozyme was crystallized from ammonium sulfate over the pH range 4.0-7.8, with protein concentrations from 100 to 150 mg ml(-1). Crystals were obtained by vapor-diffusion or batch-crystallization methods. The protein crystallized in two morphologies with an apparent morphology dependence on temperature and protein concentration. In general, tetragonal crystals could be grown by lowering the protein concentration or temperature. Increasing the temperature or protein concentration resulted in the growth of orthorhombic crystals. Representative crystals of each morphology were selected for X-ray analysis. The tetragonal crystals belonged to the P4(3)2(1)2 space group with crystals grown at pH 4.4 having unit-cell dimensions of a = b = 78.71, c = 38.6 Å and diffracting to beyond 2.0 Å. The orthorhombic crystals, grown at pH 4.8, were of space group P2(1)2(1)2 and had unit-cell dimensions of a = 30.51, b = 56.51 and c = 73.62 Å.
Crystallization of biological molecules in microgravity. Snell, EH, Chayen, NE and Helliwell, JR. The Biochemist, Vol 21, 6, 19-24 (1999).
Protein crystal movements and fluid flows during microgravity growth. Boggon, TJ, Chayen, NE, Snell, EH, Dong, J, Lautenschlager, P, Potthast, L, Siddons, DP, Stojanoff, V, Gordon, E, Thompson, AW, Zagalsky, PF, Bi, R-C, and Helliwell, JR. Phil. Trans. R. Soc. Lond. A.356, 1045-1061 (1998).
The growth of protein crystals suitable for X-ray crystal structure analysis is an important topic. The methods of protein crystal growth are under increasing study whereby different methods are being compared via diagnostic monitoring including Charge Coupled Device (CCD) video and interferometry. The quality (perfection) of protein crystals is now being evaluated by mosaicity analysis (rocking curves) and X-ray topographic images as well as the diffraction resolution limit and overall data quality. Choice of a liquid-liquid linear crystal growth geometry and microgravity can yield a spatial stability of growing crystals and fluid, as seen in protein crystallization experiments on the unmanned platform EURICA. A review is given here of existing results and experience over several microgravity missions. The results include CCD video as well as interferometry during the mission, followed, on return to earth, by rocking curve experiments and full X-ray data collection on LMS and earth control lysozyme crystals. Diffraction data recorded from LMS and ground control apocrustacyanin C(sub 1) crystals are also described.
Quality evaluation of macromolecular crystals using X-ray mosaicity measurements. Snell, E.H. Proceedings of the Montreal Spacebound 1997 meeting, Canadian Space Agency, 306-315. (1998).
Stationary crystal diffraction with a monochromatic convergent X-ray source and application for macromolecular crystal data collection. Ho JX, Snell EH, Sisk RC, Ruble JR, Carter DC, Owens SM, Gibson WM. Acta Crystallogr D Biol Crystallogr. 1998 Mar 1;54(Pt 2):200-14.
A diffraction geometry utilizing convergent X-rays from a polycapillary optic incident on a stationary crystal is described. A mathematical simulation of the resulting diffraction pattern (in terms of spot shape, position and intensity) is presented along with preliminary experimental results recorded from a lysozyme crystal. The effective source coverage factor is introduced to bring the reflection intensities onto the same scale. The feasibility of its application to macromolecular crystal data collection is discussed.
CCD video observation of microgravity crystallization of lysozyme and correlation with accelerometer data. Snell EH, Boggon TJ, Helliwell JR, Moskowitz ME, Nadarajah A. Acta Crystallogr D Biol Crystallogr. 1997 Nov 1;53(Pt 6):747-55.
Lysozyme has been crystallized using the ESA Advanced Protein Crystallization Facility onboard the NASA Space Shuttle Orbiter during the IML-2 mission. CCD video monitoring was used to follow the crystallization process and evaluate the growth rate. During the mission some tetragonal crystals were observed moving over distances of up to 200 micrometers. This was correlated with microgravity disturbances caused by firings of vernier jets on the Orbiter. Growth-rate measurement of a stationary crystal (which had nucleated on the growth reactor wall) showed spurts and lulls correlated with an onboard activity: astronaut exercise. The stepped growth rates may be responsible for the residual mosaic block structure seen in crystal mosaicity and topography measurements.
Partial improvement of crystal quality for microgravity-grown apocrustacyanin C1. Snell EH, Cassetta A, Helliwell JR, Boggon TJ, Chayen NE, Weckert E, Holzer K, Schroer K, Gordon EJ, Zagalsky PF. Acta Crystallogr D Biol Crystallogr. 1997 May 1;53(Pt 3):231-9.
The protein apocrustacyanin C1 has been crystallized by vapour diffusion in both microgravity (the NASA space shuttle USML-2 mission) and on the ground. Rocking width measurements were made on the crystals at the ESRF Swiss-Norwegian beamline using a high-resolution psi-circle diffractometer from the University of Karlsruhe. Crystal perfection was then evaluated, from comparison of the reflection rocking curves from a total of five crystals (three grown in microgravity and two earth controls), and by plotting mosaicity versus reflection signal/noise. Comparison was then made with previous measurements of almost ‘perfect’ lysozyme crystals grown aboard IML-2 and Spacehab-1 and reported by Snell et al. [Snell, Weisgerber, Helliwell, Weckert, Holzer & Schroer (1995). Acta Cryst. D51, 1099-1102]. Overall, the best diffraction-quality apocrustacyanin C1 crystal was microgravity grown, but one earth-grown crystal was as good as one of the other microgravity-grown crystals. The remaining two crystals (one from microgravity and one from earth) were poorer than the other three and of fairly equal quality. Crystal movement during growth in microgravity, resulting from the use of vapour-diffusion geometry, may be the cause of not realising the ‘theoretical’ limit of perfect protein crystal quality.
CCD video observation of microgravity crystallization: apocrustacyanin C1. Chayen, NE, Snell, EH, Helliwell, JR and Zagalsky, PF. J. Cryst. Growth 171, 219-225 (1997).
Apocrustacyanin C1 has been crystallized in the vapour-diffusion apparatus of ESA’s Advanced Protein Crystallization Facility (APCF) on-board the NASA space shuttle STS-65 International Microgravity Laboratory-2 (IML-2) mission. Crystal growth was monitored by black and white CCD observation at time intervals throughout the experiment. The resulting crystals displayed a motion within the hanging drop that is attributed to Marangoni convection effects. The images also show a “halo” effect around the growing crystals which can be attributed to the presence of depletion zones i.e. solution regions which are depleted of this coloured protein.
Lysozyme crystal growth kinetics monitored using a Mach-Zehnder interferometer. Snell EH, Helliwell JR, Boggon TJ, Lautenschlager P, Potthast L. Acta Crystallogr D Biol Crystallogr. 1996 May 1;52(Pt 3):529-33.
A Mach-Zehnder interferometer has been developed for the monitoring of the kinetics of the diffusion process in protein crystal growth. This device can be used in conjunction with the ESA Advanced Protein Crystallization Facility (APCF), which allows experiments under microgravity conditions (e.g. on board the NASA Space Shuttle). Experimental trials on the ground have been carried out with the interferometer using the engineering model of the APCF and a protein dialysis reactor. Chicken egg-white lysozyme crystal growth, as a test, has thereby been monitored directly. The changes of concentration in the solution over time have been determined via the refractive index measurements made and subsequently correlated with visual monitoring of crystal growth in a repeat experiment.
An Investigation of the perfection of lysozyme protein crystals grown in microgravity and on earth. Helliwell, JR, Snell, EH, & Weisgerber, S. Materials and Fluids Under Low Gravity. Springer Lecture notes in Physics. Vol 464, Ch. 30 edited by Ratke, L., Walter, H. & Feuerbache, B. Springer Verlag, 155-170 (1996).
Lysozyme has been used to investigate the effect of microgravity crystallisation on protein crystal perfection. Crystals were grown in the European Space Agency’s Advanced Protein Crystallisation Facility onboard the NASA Space Shuttle. Two missions of differing duration took place, Spacehab-1 and IML-2. The microgravity crystallisation time in each was 7 days and 10 hours, and 12 days and 11 hours respectively. The IML-2 crystals had grown much larger than the Spacehab-1 crystals (2.5 mm versus 0.8 mm at maximum). The earth grown control crystals, in each case, reached a size of 0.8 mm at maximum. The perfection of the crystals was evaluated with collimated, intense, synchrotron radiation. This was done using the Laue method, via the spot size, and by monochromatic rocking widths directly. For the Spacehab-1 crystals spot size measurements were carried out on station 9.5 of the SRS, along with an analysis of intensity to sigma ratio, immediately after the mission. Five months later rocking widths were measured at LURE. The IML-2 crystals were evaluated at the ESRF, on BL3 three months after their return to earth and also a further three months later on the joint Swiss-Norwegian beamline. Both the Spacehab-1 and IML-2 crystals were of exceptional perfection with the crystal mosaicity reaching values as small as 0.0010° and 0.0017° respectively. Earth-grown control crystals had values as small as 0.0032° and 0.007° respectively. There is no evidence of ‘shelf-life’ ageing of the crystals, at least over a period of 6 months, since there is close agreement of the mosaicity values from the Spacehab-1 crystals tested within weeks of that mission and the IML-2 crystals tested 6 months after that mission. The perfect mosaic block size has evidently increased over that realised in the earth-grown controls.
X-ray topography: An old technique with a new application. Stojanoff, V, Siddons, DP, Snell, EH and Helliwell, JR. Synchrotron Radiation News 9, 25-26 (1996).
The first application of X-ray topography to protein crystals.
Trends and challenges in experimental macromolecular crystallography. Chayen NE, Boggon TJ, Cassetta A, Deacon A, Gleichmann T, Habash J, Harrop SJ, Helliwell JR, Nieh YP, Peterson MR, Raftery J, Snell EH, Hädener A, Niemann AC, Siddons DP, Stojanoff V, Thompson AW, Ursby T, Wulff M. Q Rev Biophys. 1996.
Macromolecular X-ray crystallography underpins the vigorous field of structural molecular biology having yielded many protein, nucleic acid and virus structures in fine detail. The understanding of the recognition by these macromolecules, as receptors, of their cognate ligands involves the detailed study of the structural chemistry of their molecular interactions. Also these structural details underpin the rational design of novel inhibitors in modern drug discovery in the pharmaceutical industry. Moreover, from such structures the functional details can be inferred, such as the biological chemistry of enzyme reactivity. There is then a vast number and range of types of biological macromolecules that potentially could be studied. The completion of the protein primary sequencing of the yeast genome, and the human genome sequencing project comprising some 105 proteins that is underway, raises expectations for equivalent three dimensional structural databases.
Uses of the Synchrotron X-Ray Laue Technique and Investigation of Microgravity Crystal Growth, Snell EH, PhD Thesis, University of Manchester, 1995.
Dr. Snell’s PhD Thesis in Prof. John Helliwell’s group at the University of Manchester, UK.
In the first part, the Laue method was used for a structural solution of a small inorganic test crystal, NiAPO. An image plate was used for the data collection and comparison made with the use of photographic film. Synchrotron data collection was completed in less than one hour and a structure produced with an R-factor on F of 5.11% for I > 3a(I) comparing well with monochromatic studies on the same crystal. Secondly the problem of completeness of data at low resolution, “the low resolution hole” , was tackled for the protein lysozyme via a strategy involving small angular interval data collection. From a data set of 31 images recorded at 2° intervals a subset of 4°, 10° and 30° intervals (16, 6 and 3 images respectively) were compared in terms of completeness and electron density maps. The 2° data gave the best completeness of 86.1% for dmin-oo, 88.1% for dmin-2dmin and 68.6% for 2dmjn-oo and an overall improved electron density. The study documents the progressive deterioration of completeness and map quality as coarser angular intervals are employed. Thirdly the Laue method is used to examine the performance of the European Synchrotron Radiation Facility’s (ESRF) Image intensifying Charge Coupled Device (CCD) X-ray detector, with a view to helping to realise optimal detector calibration and use.
In the second part, lysozyme crystals have been grown using the European Space Agency’s (ESA) Advanced Protein Crystallization Facility (APCF) on-board the NASA STS-65 International Microgravity Laboratory-2 (IML-2) Space Shuttle mission. Exceptionally large, well defined morphology, tetragonal crystals were produced. The crystals had a 1.8 mm average length (2.5 mm maximum) along the longest dimension as opposed to 0.6 mm average (0,8 mm maximum) for the ground-control crystals grown under identical conditions (but not within an APCF). An unusual presence of orthorhombic ‘needle shaped’ crystals was also observed in the microgravity-grown case. Rocking width measurements were made at the ESRF, with a high resolution diffractometer, of a microgravity-grown and an earth-grown control, tetragonal crystal. The microgravity-grown crystal displayed mosaic spreads as low as 0.0019° compared to the lowest value of 0.0067° for the earth-grown control crystal. The reduction in mosaic spread results in a corresponding increase in peak intensity for the reflections giving improved signal to noise for microgravity-grown crystal X-ray data (Snell et al 1995, Acta Cryst. D).220 It was also readily possible to measure diffraction at 1.2 A for the microgravity-grown crystal. Secondly, a topographic study of the microgravitygrown crystals was conducted at the National Synchrotron Light Source (NSLS). Images of mosaic blocks in the crystals were obtained, the first direct evidence for these ever obtained as far as we know. The sizes of the blocks correlates well with the IML-2 grown ESRF rocking width measurements. Thirdly, a potential diagnostic tool, the Mach-Zehnder interferometer (implemented in the engineering model of the Advanced Protein Crystallization Facility) has been investigated with lysozyme as the test sample. This allowed the time-dependence of the lysozyme crystal growth process to be monitored, via the refractive index changes in the solution.
Improvements in lysozyme protein crystal perfection through microgravity growth. Snell EH, Weisgerber S, Helliwell JR, Hölzer K, Schroer K. Acta Crystallogr D Biol Crystallogr. 1995 Nov 1;51(Pt 6):1099-102.
The seminal paper discovering how a reduced acceleration growth environment improves long-range order decreasing mosaicity and enhancing signal-to-noise for the resulting reflections.
Microgravity offers an environment for protein crystallization where there is an absence of convection and sedimentation. We have investigated the effect of microgravity conditions on the perfection of protein crystals. The quality of crystals for X-ray diffraction studies is characterized by a number of factors, namely size, mosaicity and the resolution limit. By using tetragonal lysozyme crystals as a test case we show, with crystal growth in two separate Space Shuttle missions, that the mosaicity is improved by a factor of three to four over earth-grown ground control values. These microgravity-grown protein crystals are then essentially perfect diffraction gratings. As a result the peak to background of individual X-ray diffraction reflections is enhanced by a similar factor to the reduction in the mosaicity. This then offers a particularly important opportunity for improving the measurement of weak reflections such as occur at high diffraction resolution. These microgravity results set a benchmark for all future microgravity and earth-based protein crystallography procedures.
Time resolved biological and pertubation chemical crystallography: Laue and monochromatic developments. Bradbrook, G, Deacon, A, Habash, J, Helliwell, JR, Helliwell, M, Nieh, YP, Snell, EH, Trapini, S, Thompson, AW, Campbell, JW, Allinson, NM, Moon, K, Ursby, T, and Wulff, M. SPIE 2521, 160-177 (1995).
An invited paper.
Time-resolved macromolecular X-ray crystallography is a new capability for structural analysis driven by continuing improvements in synchrotron X-ray sources, optics and detectors (image plates and CCDs). Protein crystal Laue data (stationary crystal and polychromatic X-rays) were recorded at SRS Daresbury station 9.5 and ESRF Grenoble beamline 3, and processed with the Daresbury Laue software package. The Laue method allows exposure times set by the synchrotron electron bunch width, eg. 50 picoseconds. The instruments and methods developments widen opportunities for perturbation chemical crystallography studies too. A temperature dependent phase transition of a liquid crystal nickel-octahexylphthalocyanine is studied with a rapid readout CCDdetector. Structure solution by molecular replacement methods with Laue data is reported for orthorhombic lysozyme. By use of tetragonal lysozyrne as a test case it is shown that with fine angular intervals, wide total angular coverage of Laue exposures and the deconvolution of multiples, good connectivity of electron density maps can be realised. The monochromatic rotating crystal method offers possibilities of extremely fast rotations which allow a complete data set to be recorded onto a single image -Large-angle oscillation technique (LOT). The processed LOT data looks promising. LOT electron density maps are presented. We are now exploring the use of 3600 rotations onto a single image. However, since the time required to cover one revolution of the crystal is likely to be greater than 1 millisecond, and because the monochromatic synchrotron radiation beam intensitysets its own limits to the shortest exposure time, the Lane method will retain its role for the fastest exposures incrystallography. Laue techniques open up quicker data collection opportunities in neutron protein crystallography
Image-plate synchrotron laue data collection and subsequent structural analysis of a small test crystal of a nickel-containing aluminophosphate. Snell E, Habash J, Helliwell M, Helliwell JR, Raftery J, Kaucic V, Campbell JW. J Synchrotron Radiat. 1995 Jan 1;2(Pt 1):22-6.
The first use of an image plate with Laue data for a small molecule structure.
Image plates have advantages over photographic films, which include wider dynamic range, higher detector quantum efficiency, reduced exposure time and large size. In this study, an on-line image-plate system has been used to record crystallographic data from a small crystal. In particular, synchrotron Laue data were recorded with lambda(min) = 0.455, lambda(max) = 1.180 A, in 20 images 10 degrees apart and with an exposure time of 0.3 s each from a crystal (0.02 x 0.05 x 0.25 mm) of a nickel-containing aluminophosphate, NiAPO. The Laue data were analyzed with the Daresbury Laue software, including the application of an absorption correction. The structure was solved by a combination of the Patterson method and successive difference Fourier calculations using SHELXS86 and SHELXL93; the final R value for 1934 unique reflections (all data) and 310 parameters was 7.90%. The structure agrees with that determined by monochromatic diffractometry using the same crystal and reported by Helliwell, Gallois, Kariuki, Kaucic & Helliwell [Acta Cryst. (1993), B49, 420-428] with an r.m.s. deviation of 0.03 A. Hence, this study shows the image-plate device to be very effective for synchrotron data collection and subsequent structure analysis from small crystals, i.e. 0.02 x 0.05 x 0.25 mm, in chemical crystallography as well as providing further confirmation of the practicability of Laue data in structure solution and refinement..
Electron density maps of lysozyme calculated using synchrotron Laue data comprising singles and deconvoluted multiples. Campbell , JW, deacon, A, Habash, J, Helliwell, JR, McSweeney, S, Quan, H, Raftery, J and Snell, E. Bull. Mater. Sci., 17,1, 1-18 (1994).
We have used lysozyme as a test case to illustrate the application of a new method of estimating the intensities of Laue multiples reflection data in protein crystallography. Hen egg-white lysozyme, an enzyme with a single polypeptide chain of 129 amino acids, crystallizes in space group P43212 with cell parametersa=b=79·1 Å,c=37·9 Å. Laue image plate data were collected using synchrotron radiation on station 9·5 at Daresbury with a total exposure time of 0·95 seconds. The data processed were separated into two data sets comprising the singles and the combined singles and deconvoluted multiples. The method of deconvolution was that of Campbell and Hao (1993), which utilizes the intensity variation of theλ-curve. Electron density maps (2F o−F c) based on the two data sets are then compared. This comparison shows that the deconvoluted multiples do indeed contribute usefully to the continuity of the maps due to the improved completeness of the data. A number of map sections along the polypeptide chain, based on the two Laue data sets, are shown for comparison. These include Arg 5, His 15, Phe 38, Asp 52, Tyr 53, Pro 70, Trp 108 and the four disulphide bridges.
The emergence of the synchrotron Laue method for rapid data collection from protein crystals. Cassetta, A, Deacon, A, Emmerich, C, Habash, J, Helliwell, JR, McSweeney, S, Snell, E, Thompson, AW and Weisgerber, S. Proc. R. Soc. Lond. A, 177-192 (1993).
Synchrotron X-radiation (SR) is intense, polychromatic and collimated. It is widely exploited, in macromolecular crystallography, particularly using a monochromatized short wavelength beam. The spectral curve of SR, however, ideally lends itself to use of Laue geometry, i.e. the original diffraction experimental arrangement based on a stationary crystal and a polychromatic X-ray beam. Rapid exposure times and time-resolved crystallography studies, e.g. of enzymes, are now possible. Historical objections to the use of Laue diffraction data, particularly the multiplicity distribution, have been found not to be as limiting as once thought. The credentials of the Laue method have been established through a variety of Laue crystal structure analyses, involving photographic film as detector. Recently a three-dimensional arrangement of films, known as a toast-rack, has been used to alleviate problems with spatially overlapping spots. This paper provides a review of these results and then reports several developments. In particular, one of the first Laue analyses using an image plate as detector, namely of a cobalt substituted concanavalin A crystal, is discussed. Recent experimental developments, also at the Daresbury synchrotron, are then described. First, a large toast-rack has been used to record Laue data from a protein crystal. Secondly, a transmission X-ray mirror has been constructed from thin mylar (1.5 μm) and used to provide a λ max filter instead of using aluminium foils. Thirdly, since the Laue method suffers from poor sampling of the low resolution data, a new method (known as LOT) has been introduced.